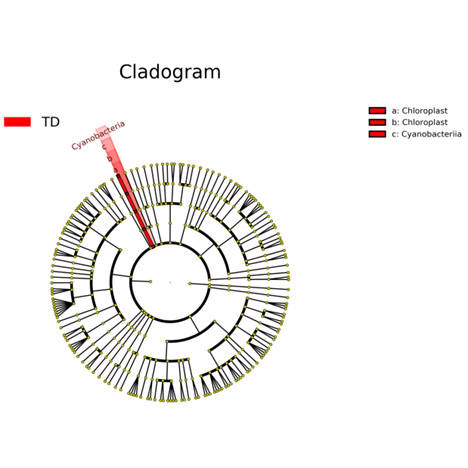
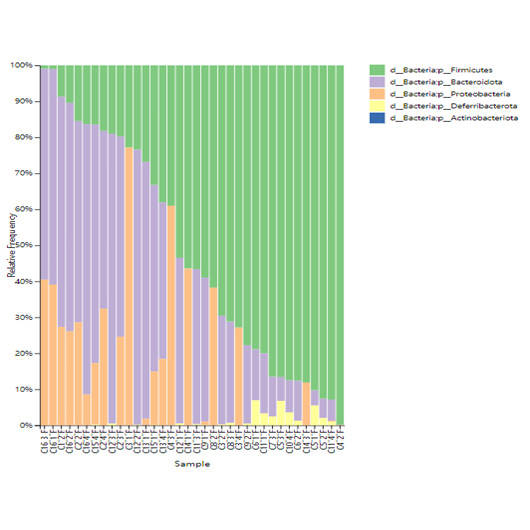
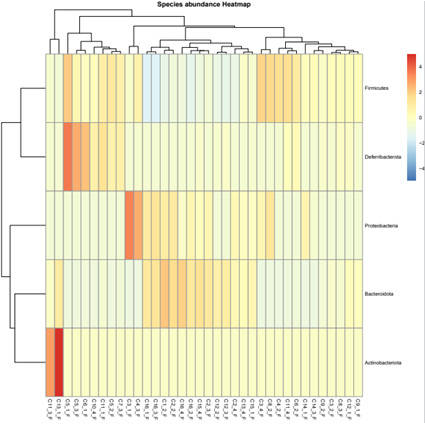
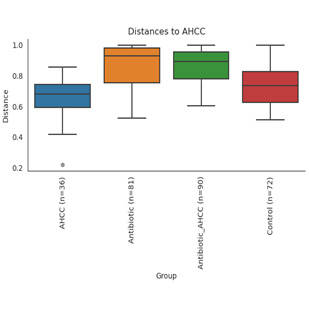
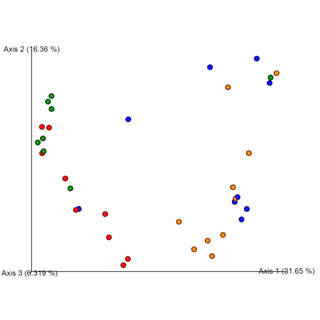
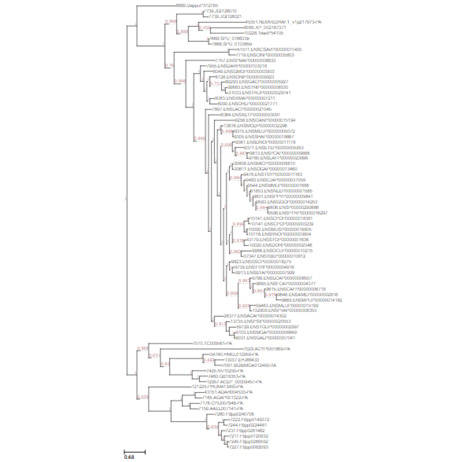
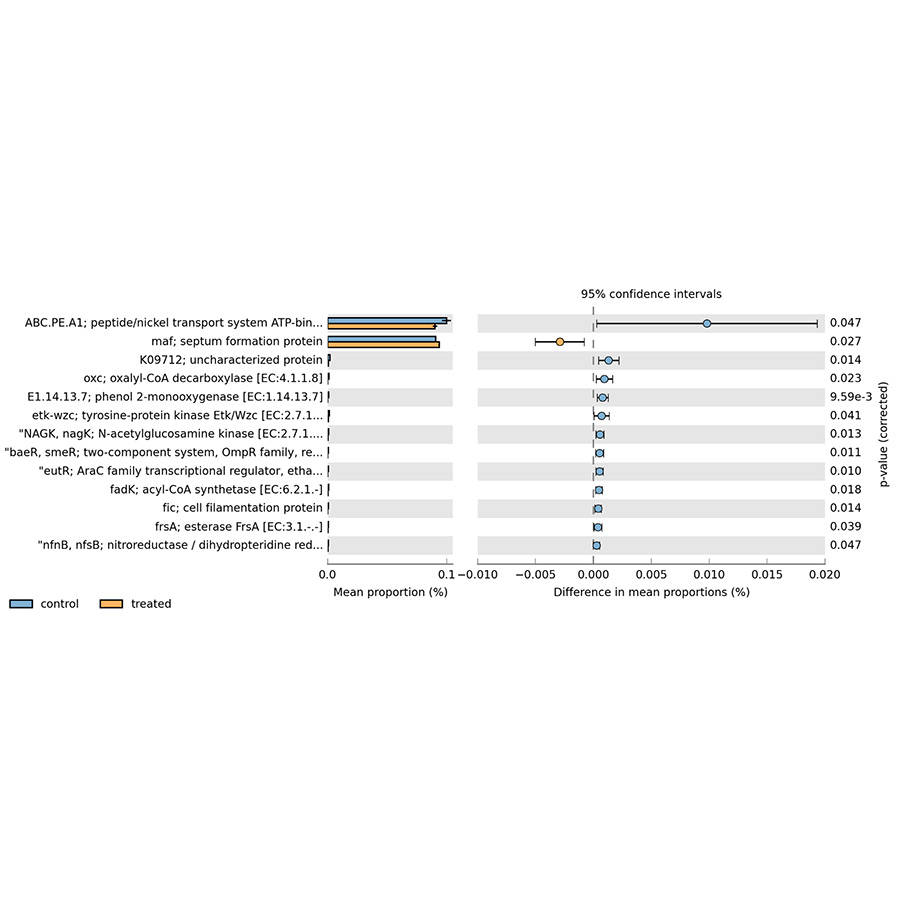
Prokaryotes, encompassing bacteria and archaea, are unicellular entities devoid of a nucleus. CD Genomics provides sophisticated sequencing solutions targeting Bacteria, Archaea, and Actinomycetes. Our services are designed to decode their genetic architectures and scrutinize microbial interactions within various ecosystems.
Eukaryotes, characterized by cells with a nucleus, include organisms such as fungi and yeast. CD Genomics delivers comprehensive sequencing for Fungi and Yeast, facilitating an in-depth examination of their genetic diversity and ecological roles. Our sequencing data elucidates patterns in their genomic structures and evolutionary histories.
Viruses are genetic elements that depend on host cells for replication. CD Genomics specializes in sequencing Bacteriophages as well as a broad spectrum of viruses. Through meticulous genome mapping, we advance research in viral biology, genetic variability, and the impacts on host populations.