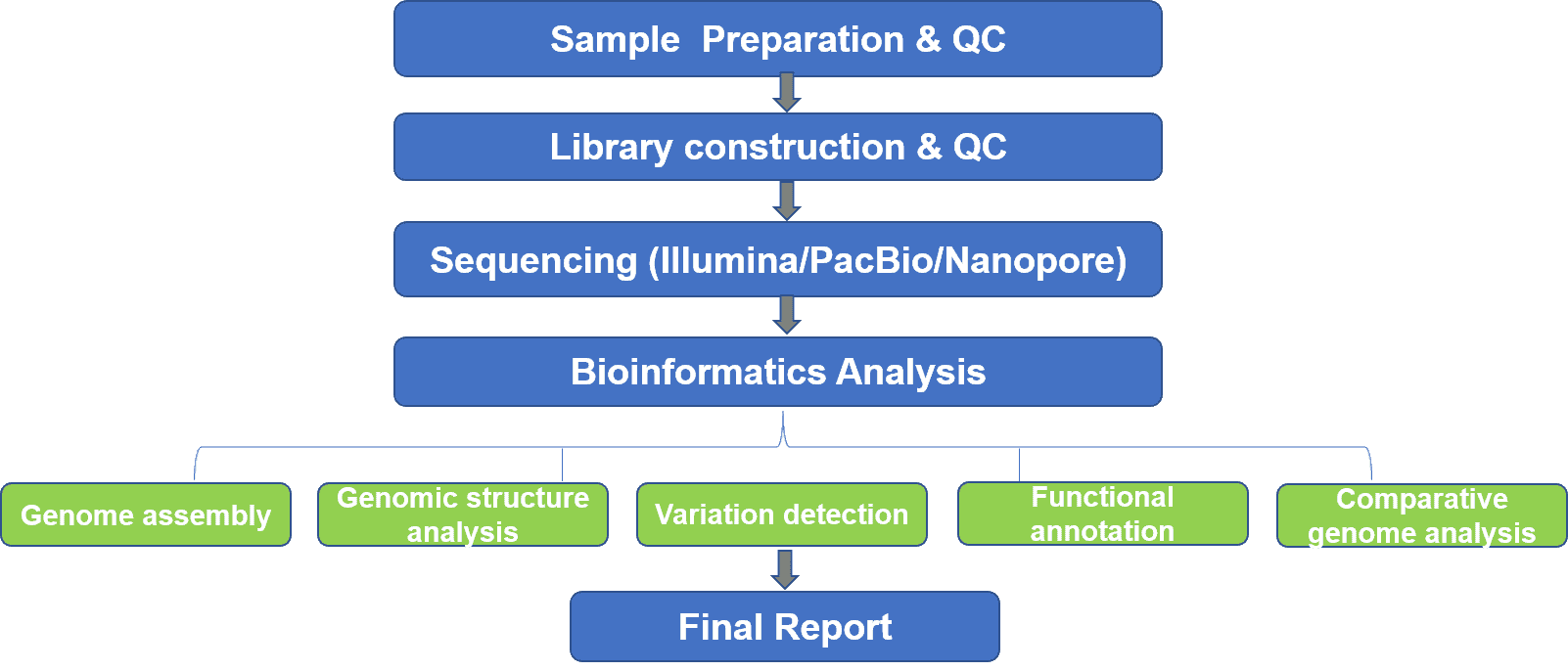
Yeast whole genome sequencing sequences the entire yeast genome in one run, including chromosomal DNA as well as mitochondrial DNA. We utilize both NGS and long-read sequencing platform to provide whole-genome de novo sequencing and re-sequencing of yeasts. De novo sequencing refers to sequencing a novel genome without reference sequence. The coverage quality of de novo assembly predominantly depends on the size and continuity of the contigs. Long-read sequencing including both Oxford nanopore and PacBio sequencing systems enables high-quality genome assembly with improved continuity as the contig N50 length is ten times longer than an NGS-only assembly. Highly accurate NGS data can be used to polish the genome assembly.
While yeast whole-genome de novo sequencing can be used to reconstruct the first genome map for yeast species, a valuable reference genome for re-sequencing and whole-genome resequencing is commonly required to identify variations such as SNPs, InDels, SVs, CNVs, and other genetic changes, which may be potential biomarkers. Yeast whole genome sequencing provides an unparalleled opportunity to characterize the polymorphic variants in a yeast population, which comprehensively unravels the underlying mechanisms of origin, development, growth, and evolution of yeasts.