Microbial Diversity Analysis – 16S/18S/ITS Sequencing
- Overview
- Service Specifications
- FAQs
- Demo
- Customer Case
Overview
Our microbial diversity analysis platform, leveraging 16S/18S/ITS amplicon sequencing, is designed to characterize microbial diversity with heightened efficiency and cost-effectiveness. By incorporating next-generation and third-generation sequencing technologies, along with supplementary molecular techniques, we ensure comprehensive and precise analyses. Our team of scientists employs this cutting-edge platform to support our clients in fulfilling both scientific research and regulatory compliance mandates.
Our Advantages:
- Comprehensive Insights: Advanced 16S, 18S, and ITS sequencing for detailed analysis of prokaryotic and eukaryotic communities.
- High Sensitivity: Detect microbial taxa at abundances as low as 0.01% for precise identification.
- Rapid Results: Expedite your research with prompt findings.
- Expert Support: Access guidance from experienced scientists for optimal data interpretation.
- Flexible Platforms: Utilize various sequencing technologies tailored to your research needs.
- Diverse Analysis: Assess a wide range of microbes, including bacteria, fungi, and actinobacteria.
Why is Microbial Diversity Analysis Important
The analysis of microbial diversity is of paramount importance across several disciplines, including environmental science, agriculture, and human health. A rich diversity in microbial communities underpins ecosystem stability, facilitates nutrient cycling, and enhances disease resistance. For instance, within soil ecosystems, a varied microbiome can significantly promote plant growth and increase resilience against pathogens. In the context of human health, the diversity of gut microbiota is intricately linked to metabolic health and immune function. Through the assessment of microbial diversity, researchers can identify shifts that may indicate ecological disturbances or potential health issues, thereby enabling timely and effective interventions.
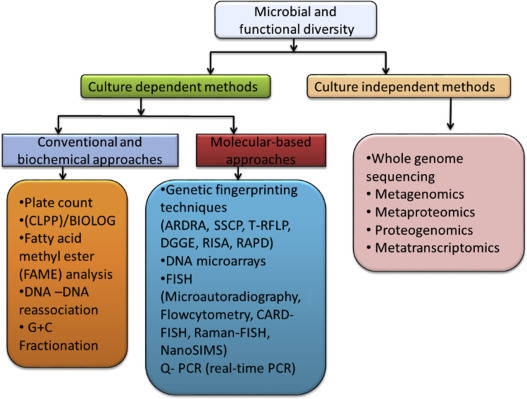
How Do You Analyze Microbial Diversity
The analysis of microbial diversity is crucial for elucidating ecosystem dynamics and the ecological roles of microorganisms. This requires high-resolution sequencing methods targeting specific genetic regions such as 16S rRNA, 18S rRNA, and Internal Transcribed Spacer (ITS). The primary approaches in this field include second-generation sequencing, third-generation sequencing, and metagenomic sequencing.
Second-Generation Sequencing
Second-generation sequencing, often utilizing Illumina technology, targets genetic regions within 16S rRNA, 18S rRNA, and ITS. This high-throughput approach permits the simultaneous detection of dominant, rare, and novel species within a sample. By amplifying and sequencing specific variable regions, researchers gain crucial insights into the composition and relative abundances of microbial communities, thereby elucidating ecological interactions with greater detail.
Third-Generation Sequencing
Conversely, third-generation sequencing platforms like PacBio leverage Single Molecule Real-Time (SMRT) technology to achieve full-length sequencing of target genes. This method emphasizes accuracy and fidelity, providing a more comprehensive overview of microbial community structures. It captures the complete variability of sequences, thus facilitating precise phylogenetic analyses.
Metagenomic sequencing extends beyond targeted sequencing by analyzing entire microbial genomes directly from environmental samples. This technique not only profiles microbial diversity but also uncovers functional traits, inter-species relationships, and environmental interactions. Through metagenomics, researchers can elucidate significant biological insights, thereby enhancing our understanding of complex microbial ecosystems.
Service Specifications
Introduction to Our Microbial Diversity Analysis Platform
Our microbial diversity detection platform synergizes advanced sequencing technologies with quantitative PCR (q-PCR) to facilitate precise analyses of 16S, 18S, and ITS regions. It accommodates both short-read and full-length sequencing methodologies, with results rigorously validated through Sanger sequencing or q-PCR, ensuring accurate species quantification. This platform encompasses both prokaryotic and eukaryotic diversity analyses, utilizing 16S rRNA for bacteria and archaea, and ITS and 18S rRNA for fungi and eukaryotes, thus enabling a comprehensive exploration of microbial diversity across diverse ecosystems.
Our Services
- Bacterial 16S rRNA Sequencing: This service targets bacterial diversity by amplifying and sequencing the 16S rRNA gene, offering valuable insights into the composition of bacterial communities.
- Archaeal 16S rRNA Sequencing: Focused on archaea, this sequencing reveals the diversity and ecological roles of these microorganisms within their environments.
- Diversity Analysis of Actinomycetes: This analysis assesses the diversity of actinomycetes, which are crucial for understanding ecological functions and exploring potential biotechnological applications.
- Fungal ITS Sequencing: Utilizing Internal Transcribed Spacer (ITS) sequencing, this service identifies and characterizes fungal communities present in various habitats.
- Eukaryotic 18S rRNA Sequencing: This analysis examines eukaryotic diversity by sequencing the 18S rRNA gene, providing a comprehensive overview of protists and other eukaryotic organisms.
- Full-Length 16S/18S/ITS Sequencing: This service offers complete sequencing of 16S, 18S, and ITS regions, enabling an in-depth analysis of microbial community structure and function.
- 2bRAD-M Analysis for Microbiome: Employing 2bRAD-M technology, this high-resolution analysis enhances our understanding of complex microbial interactions within microbiomes.
We also offer metagenomic sequencing; for further details, please feel free to contact us.
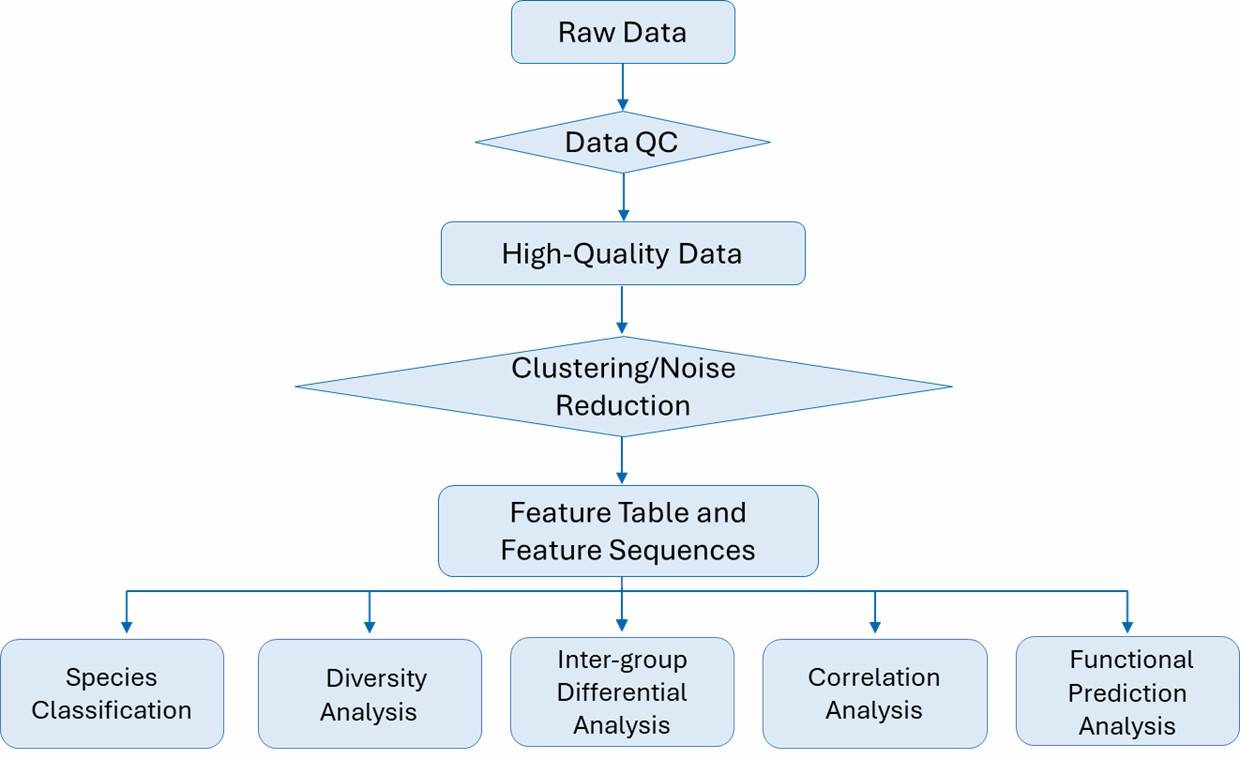
Microbial Diversity Analysis Workflow
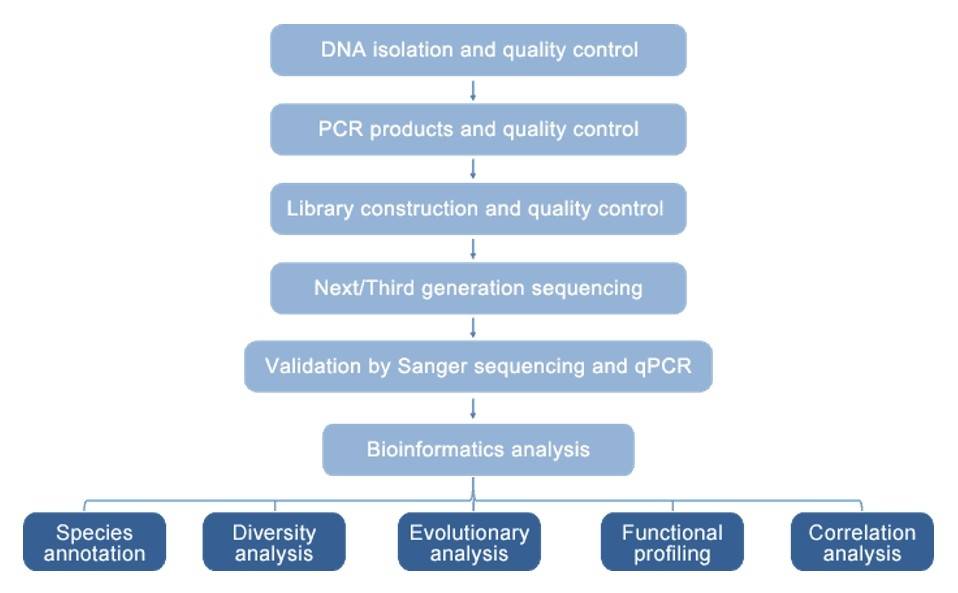
Technical Parameters
- Illumina NovaSeq, Hiseq, MiSeq; PE250
- PacBio SMRT, Library Type: 2 K, Data Volume: Approximately 3,000 to 40,000 CCS reads per sample
Bioinformatics Analysis
Our bioinformatics analysis includes five parts: species annotation, diversity analysis, evolutionary analysis, functional profiling, and correlation analysis. For advanced bioinformatics analysis in detail, please refer to the following table.
| Bioinformatics analysis |
The problem to be solved |
| Species annotation |
OTU clustering, species annotation |
| Diversity analysis |
α diversity |
Species richness within a single microbial ecosystem |
| β diversity |
Diversity in microbial community between different environments |
| Meta-analysis |
Comparative studies of abundant features between datasets |
| Evolutionary analysis |
Construction of phylogenetic tree |
| Functional analysis |
Analysis of the Intestinal Microflora |
Locate the gut bacteria, predict human disease, and provide disease treatment |
| PICRUSt |
Predict functional profiles |
| FAPROTAX |
Predict the process of biochemical cycles |
| Tax4fun |
Predict functional profiles |
| FunGuild |
Predict the ecological roles of eukaryotes |
| Correlation analysis |
CCA |
Study the relationships between microbes and environmental factors |
| VPA |
| Network analysis |
Sample Requirement
| Service |
Sample Type |
Recommended Quantity |
Minimum Quantity |
Concentration |
| 16S/18S/ITS Sequencing |
Genomic DNA |
≥100 ng |
10 ng |
≥1 ng/μl |
| Full-Length 16S/18S/ITS Amplicon Sequencing |
Genomic DNA |
≥ 500ng |
|
10 ng/µL |
| Tissue |
1-3g |
1 g |
|
| Thallus |
5 g |
3 g |
|
| Interstitial Fluid |
3-5 mL |
1 mL |
|
| Environmental Samples |
3-5g |
1 g |
|
| Water filter membrane |
3 |
1 |
|
Note: If you wish to obtain more accurate and detailed information regarding sample requirements, please feel free to contact us directly.
Deliverables
- Raw sequencing data
- Trimmed and stitched sequences (FASTA)
- Data QC report
- Bioinformatics analysis report
FAQs
Microbial Diversity Analysis FAQ
- 1. What does OTU refer to in microbial diversity?
- 2. What are the differences between the four Beta diversity distance algorithms?
- 3. What is the difference between PCA and PCoA?
Demo
Demo
Partial results of our microbial diversity analysis – 16S/18S/ITS sequencing service are shown below:
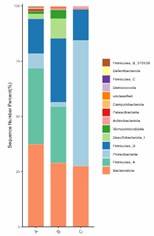
Microbial Distribution Bar Chart
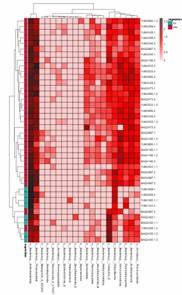
Classification Heatmap
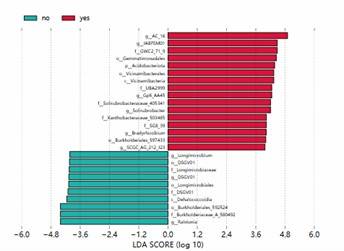
LEfSe Analysis LDA Bar Chart
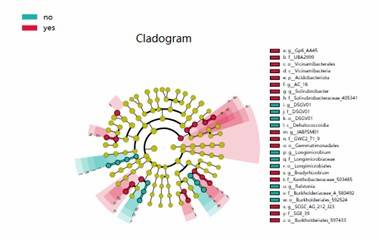
LEfSe Analysis Cladogram
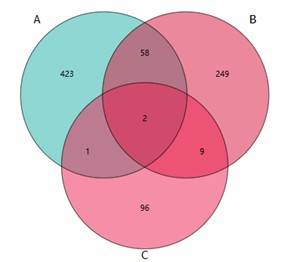
Venn Diagram
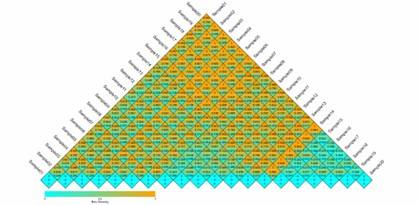
Beta Diversity Index Heatmap
Customer Case
Customer Case
Microbiota of the Digestive Gland of Red Abalone (Haliotis rufescens) Is Affected by Withering Syndrome
Journal: Microorganisms
Impact factor: 4.26
Published: 2020
Find out more
Backgrounds
The study centers on Withering Syndrome (WS), an infectious disease that heavily impacts abalone aquaculture. This disease is caused by the intracellular bacterium Candidatus Xenohaliotis californiensis, which infects the digestive glands, disrupting their function and causing progressive wilting in abalones. While the direct effects of WS are understood, its impact on the microbiota of the digestive glands in abalones is not well-studied. The main goal of this research is to determine if there are differences in the digestive gland-associated microbiota between healthy red abalones and those affected by WS.
Materials & Methods
Sample preparation:
-
Red abalone
- Converted land samples
- DNA extraction
Data Analysis:
-
Statistical analysis
- Beta diversity measures
- Community composition testing
- Differential abundance analysis
Results
The study found notable differences in microbiota composition between healthy and WS-affected abalones. Healthy abalones exhibited a diverse and balanced bacterial community, whereas WS-affected abalones had a microbiota dominated by specific bacteria associated with the disease. One of the most significant findings was that a certain bacterium was dominant in healthy abalones, while the primary pathogen of WS was more prevalent in the affected group. Interestingly, the pathogen was also present in some healthy specimens, indicating that the balance between different bacterial populations might be crucial in determining the disease status.
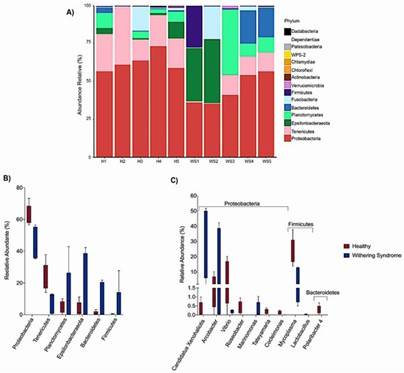 Figure 1. (A) Relative abundance of microbiota at the phylum level in the digestive glands of healthy (H1-H5) and withering syndrome-affected red abalone (WS1-WS5). (B) Comparison of relative abundance at the phylum level between healthy (red boxes) and affected (blue boxes) abalone. (C) Comparison at the genus level.
Figure 1. (A) Relative abundance of microbiota at the phylum level in the digestive glands of healthy (H1-H5) and withering syndrome-affected red abalone (WS1-WS5). (B) Comparison of relative abundance at the phylum level between healthy (red boxes) and affected (blue boxes) abalone. (C) Comparison at the genus level.
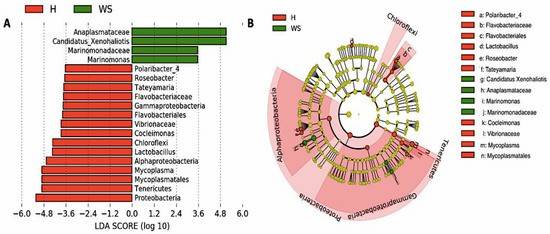 Figure 2. Differences in digestive gland microbiota of healthy red abalones (H) compared with red abalones with withering syndrome disease (WS).
Figure 2. Differences in digestive gland microbiota of healthy red abalones (H) compared with red abalones with withering syndrome disease (WS).
Conclusions
The findings suggest that WS significantly disrupts the microbiota composition in the digestive glands of red abalones. Specific bacterial communities are associated with either health or disease, highlighting the potential ecological role of microbiota in WS pathogenesis. Further research is needed to deepen our understanding of the dynamics of digestive gland microbiota and its influence on the progression of Withering Syndrome in abalones.
Reference
- Villasante A, Catalán N, Rojas R, et al. Microbiota of the digestive gland of red abalone (Haliotis rufescens) is affected by withering syndrome. Microorganisms. 2020 Sep 13;8(9):1411.
* For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.











 Figure 1. (A) Relative abundance of microbiota at the phylum level in the digestive glands of healthy (H1-H5) and withering syndrome-affected red abalone (WS1-WS5). (B) Comparison of relative abundance at the phylum level between healthy (red boxes) and affected (blue boxes) abalone. (C) Comparison at the genus level.
Figure 1. (A) Relative abundance of microbiota at the phylum level in the digestive glands of healthy (H1-H5) and withering syndrome-affected red abalone (WS1-WS5). (B) Comparison of relative abundance at the phylum level between healthy (red boxes) and affected (blue boxes) abalone. (C) Comparison at the genus level. Figure 2. Differences in digestive gland microbiota of healthy red abalones (H) compared with red abalones with withering syndrome disease (WS).
Figure 2. Differences in digestive gland microbiota of healthy red abalones (H) compared with red abalones with withering syndrome disease (WS).