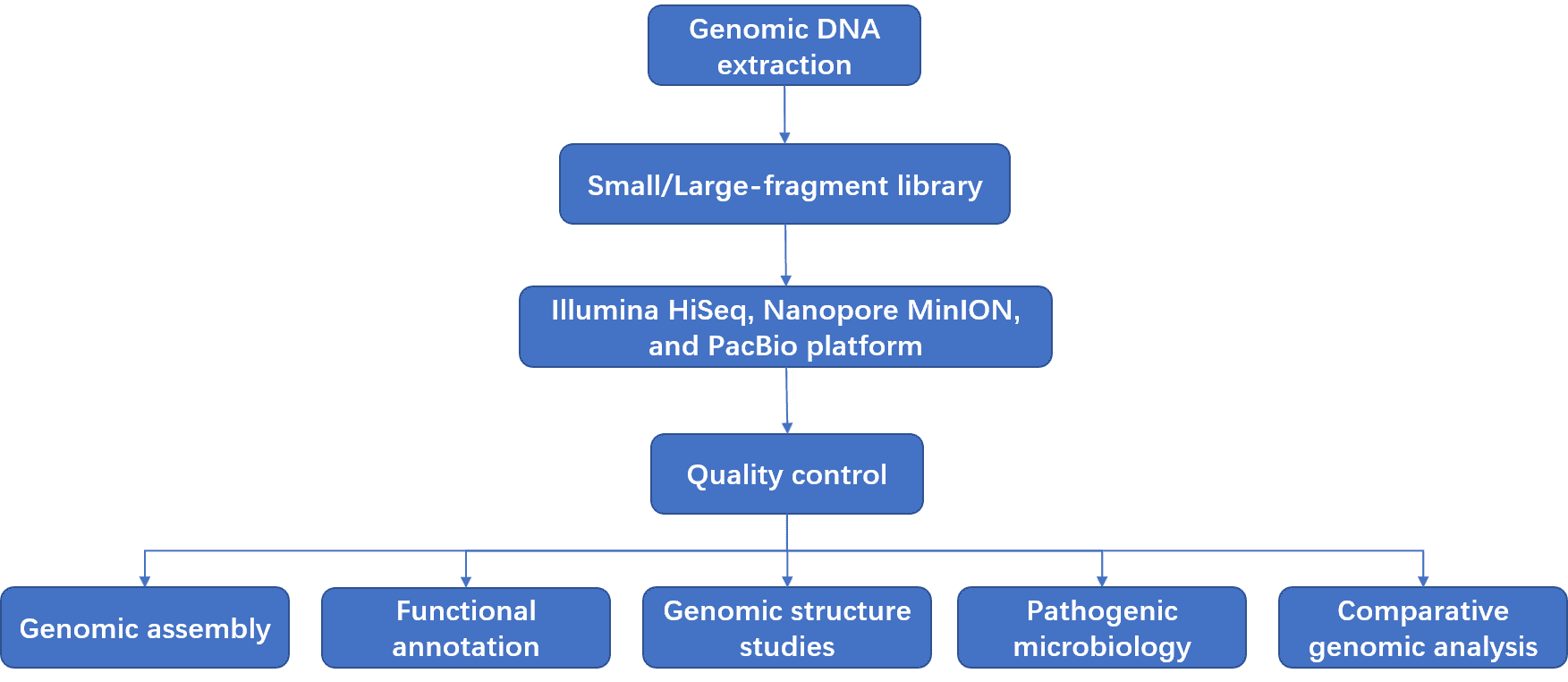
We provide next generation sequencing (NGS) and long-read sequencing technologies to sequence the fungal whole genome. Our sequencing platforms include the advanced Illumina HiSeq/MiSeq (PE150/251), Nanopore, and PacBio SMRT instruments, which can quickly and efficiently produce high-quality sequencing data. Fungal whole genome sequencing includes two categories: fungal genome de novo sequencing and fungal genome resequencing. The workflow for fungal genome de novo sequencing includes the following steps: (1) the chromosomal DNA is randomly interrupted; (2) different lengths of sequences are selected to construct a library; (3) large-scale sequencing; (4) genomic assembly without any prior genome information; (5) fill the gaps on the draft genomes. The genome sequence of fungal individuals or populations can also be determined through alignment to reference genome sequences using the homogeny of sequences as an index. It is a quick and accurate method for detection of variants.

Fungal whole genome sequencing can be used to identify pathogenic fungi, genes related to pathogen and host interaction, and the evolutionary relationships among closely related species. For edible and medicinal fungi, it can be used to discover complex metabolic pathways and to identify metabolites which are beneficial to human health. And some fungi have implications for biological control or industrial applications. Our platform provides data on the fungal genome sequence, helping to improve medical science, agriculture science, ecology, bioremediation, bioenergy, and biotech industries.