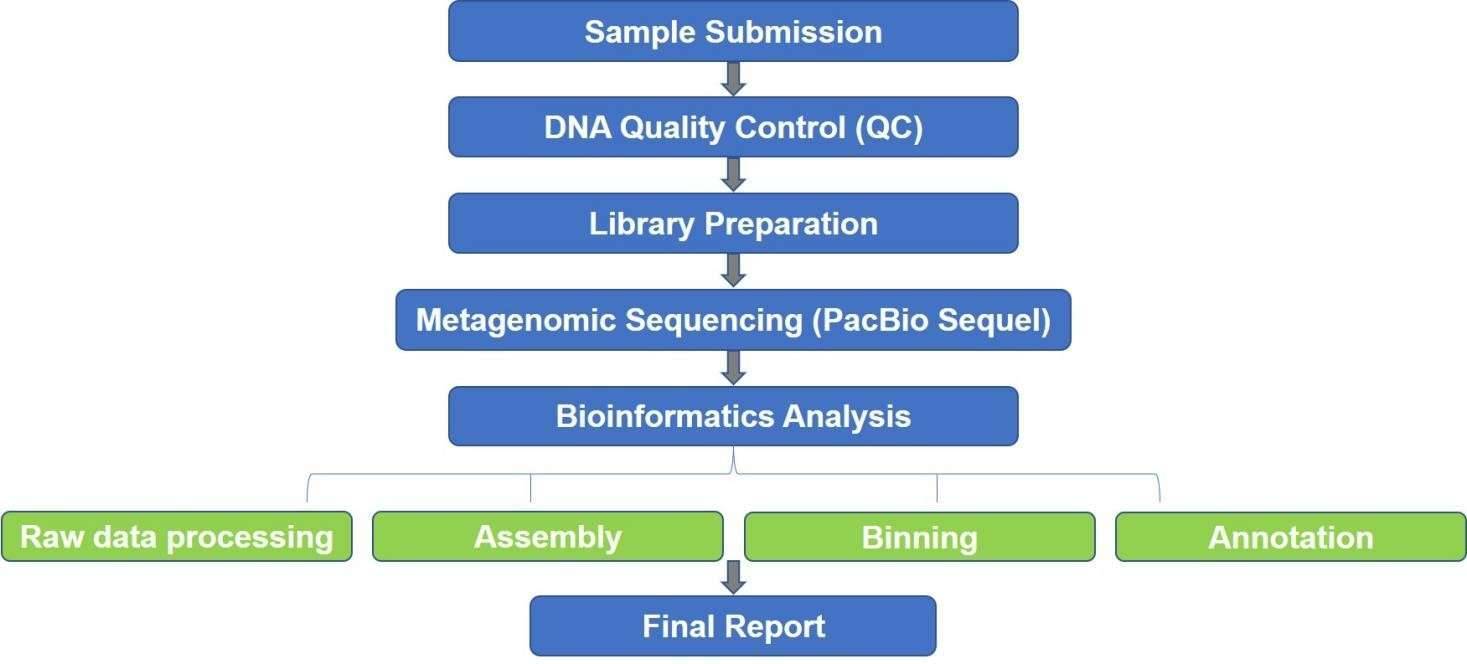
Microorganisms are almost everywhere. But to study them, more than 99% cannot be cultured in vitro. Our SMRT-based metagenomics, using PacBio RSII or Sequel from PacBio Biosciences, is a highly accurate, culture-independent metagenome profiling approach to study the microbial communities despite the culture limitations. The PacBio SMRT sequencing technology is characterized by unparalleled read length and accuracy, which is helpful for the reconstruction of the high-quality draft and metagenomic analysis.
SMRT-based metagenomics can be used to study the species composition and abundance, identify differentially expressed genes (DEGs) among samples, explore gene functions and metabolic pathways, and mine gene resources for bioactive products. The technology can also be used to elucidate and understand the relations between microbes and their habitat/host, like the microenvironments within the human body in healthy or diseased states. SMRT-based metagenomics has been widely applied to research and biotech industry.