Chromatin Remodeling Complexes: Mechanisms, Technologies, and Clinical Frontiers
In the field of genetics, the genome can be likened to a voluminous tome of life, with chromatin remodeling complexes serving as the unseen forces that meticulously manipulate its pages. These complexes, by adjusting or reorganizing the positioning of nucleosomes, determine which genes are to be "uncovered" for transcription and which remain "sealed" in their repressed state. Historically, the scientific community has primarily concentrated on the SWI/SNF and Polycomb families. However, advancements in technology have progressively unveiled the distinctive functions of additional remodeling complexes, such as NuRD and ISWI. These revelations have not only led to significant revisions in scientific literature but also have furnished novel approaches for therapeutic interventions.
Molecular Mechanisms of Chromatin Remodeling
Chromatin remodeling complexes are implicated in a variety of processes that extend far beyond the simple activation or repression of specific genes. These complexes function as precision machinery, dynamically regulating the physical state of chromosomes by synergizing multiple enzyme activities. In recent years, the combination of high-resolution imaging and epigenomics techniques has enabled scientists to delve deeper into the molecular mechanisms of these complexes.
NuRD Complex: The Dual Sided Hand of DNA Repair and Cell Differentiation
The NuRD complex is a multifaceted protein that functions in two distinct ways. Firstly, it repairs DNA damage and, secondly, it directs cell differentiation. It achieves this by recruiting histone deacetylase (HDAC), a process that involves the removal of acetyl groups from chromatin. This, in turn, creates a tighter local structure that allows for the action of DNA repair proteins, such as BRCA1. Concurrently, NuRD can also "label" histones with methyltransferase G9a, leading to the temporary shutdown of gene expression in the damaged region and the prevention of "garbled" genetic information during the repair process. In mouse embryonic stem cells, the absence of NuRD results in the accumulation of DNA damage, akin to untreated holes, eventually leading to the failure of stem cells to differentiate into nerve cells.
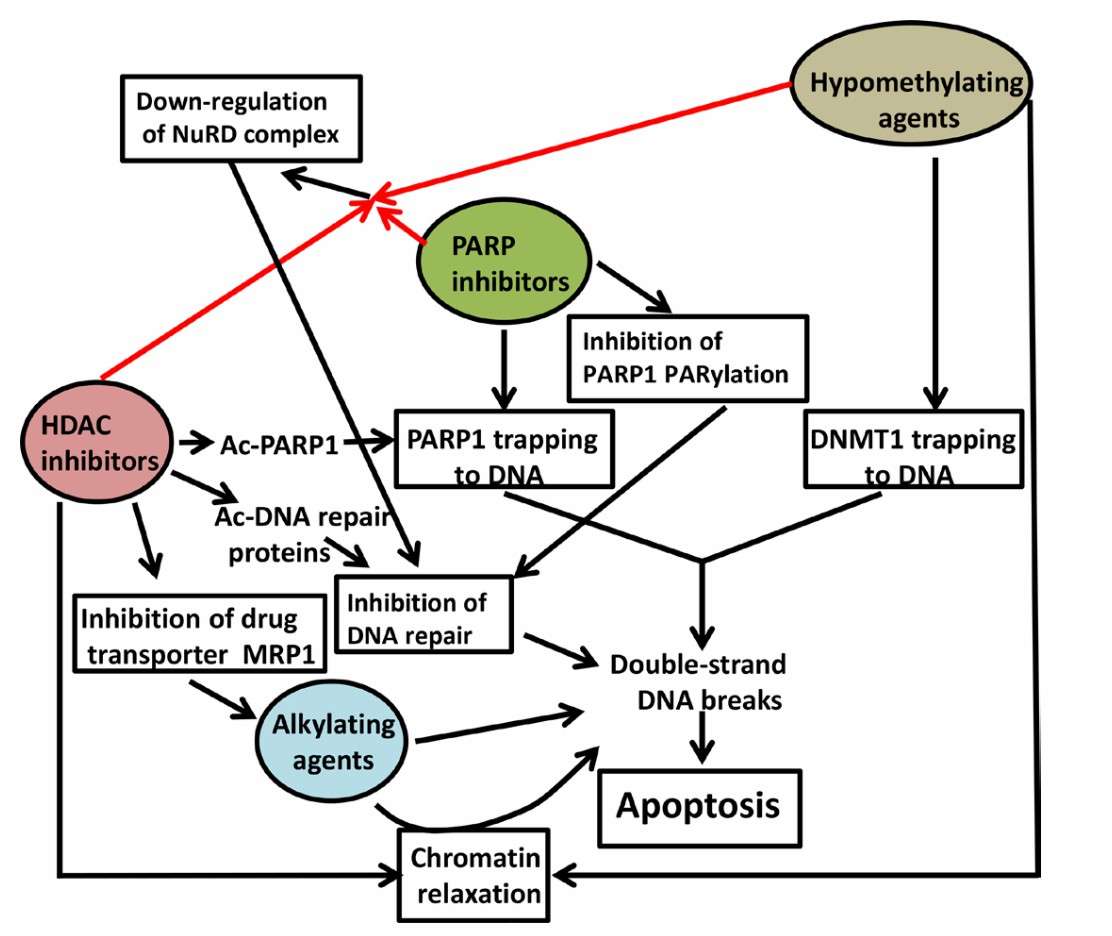 Suggested mechanisms of synergistic cytotoxicity (Valdez et al., 2017)
Suggested mechanisms of synergistic cytotoxicity (Valdez et al., 2017)
The dynamic behavior of NuRD is often captured through the use of single-cell ATAC-seq technology, a technique that utilizes high-throughput sequencing to detect open regions of chromatin. This method facilitates the identification of variations in NuRD activity across distinct cellular subpopulations. A notable illustration of this phenomenon emerges from the analysis of single-cell ATAC-seq data during the process of hematopoietic stem cell differentiation, which reveals that NuRD tends to bind to the regulatory regions of specific transcription factors, such as GATA1, and that it maintains cell-fate orientation by shutting down unrelated genes.
The ISWI family: "urban planners" of genomic space
The ISWI family of complexes (e.g., SNF2L) has been shown to play a critical role in maintaining the "spatial order" of the genome. These complexes function by sliding nucleosomes, akin to bulldozers, ensuring that the chromatin fibers remain evenly spaced. This, in turn, affects the structure of the genome in three dimensions. Recent advancements in Hi-C technology, which utilizes cross-linking DNA and high-throughput sequencing to map three-dimensional chromatin interactions, have elucidated a crucial role for ISWIs in maintaining the boundaries of topologically associated domains (TADs). The absence of ISWIs has been observed to result in the blurring of these boundaries, potentially leading to the activation of cancer-causing genes that would otherwise have been segregated. For instance, in gliomas, ISWI deficiency has been found to be closely associated with chromatin decompression in the EGFR gene region. This provides a theoretical basis for the development of anti-cancer drugs that target ISWI.
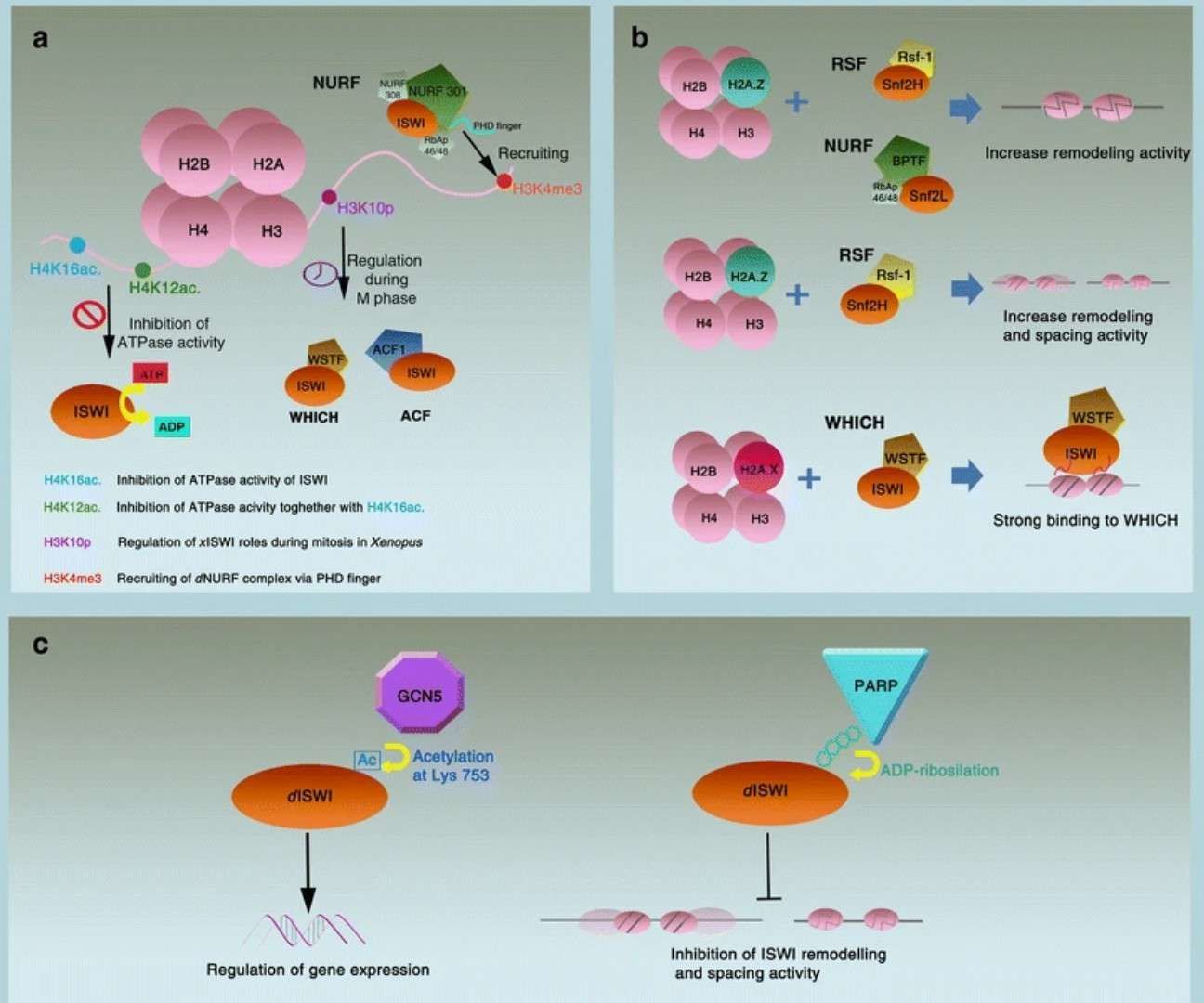 Regulation of ISWI chromatin remodellers by histones and post-translational modifications (Toto et al., 2014)
Regulation of ISWI chromatin remodellers by histones and post-translational modifications (Toto et al., 2014)
More sophisticated studies necessitate a synthesis of epigenomic peak annotation analysis. This technology can identify precise binding sites of ISWI complexes from substantial sequencing data and annotate the chromatin states (e.g., enhancers or promoters) in which these sites are situated. When integrated with an analysis of multi-omics data, scientists can construct a comprehensive regulatory network from chromatin structure to gene expression and even predict which genes may be uncontrolled due to ISWI dysregulation.
Advanced Technologies for Chromatin Dynamics
Chromatin remodeling is a dynamic process, and conventional techniques frequently only capture static snapshots. In recent years, two technological innovations have transformed the field. These innovations enable scientists to "see" the spatial connectivity of genomes in three dimensions and even track the movement of complexes in real time.
Three-dimensional genome mapping: from Hi-C to single-cell resolution
Spatial folding of chromosomes directly impacts the regulation of gene expression. Hi-C technology facilitates the identification of topologically associated domains (TADs) and enhancer-promoter loops by cross-linking DNA and conducting high-throughput sequencing to map the three-dimensional interactions of chromosomes. In Alzheimer's disease research, Hi-C data revealed an abnormal folding of the BACE1 gene region, caused by a defective ISWI complex that promotes pathological accumulation of β-amyloid.
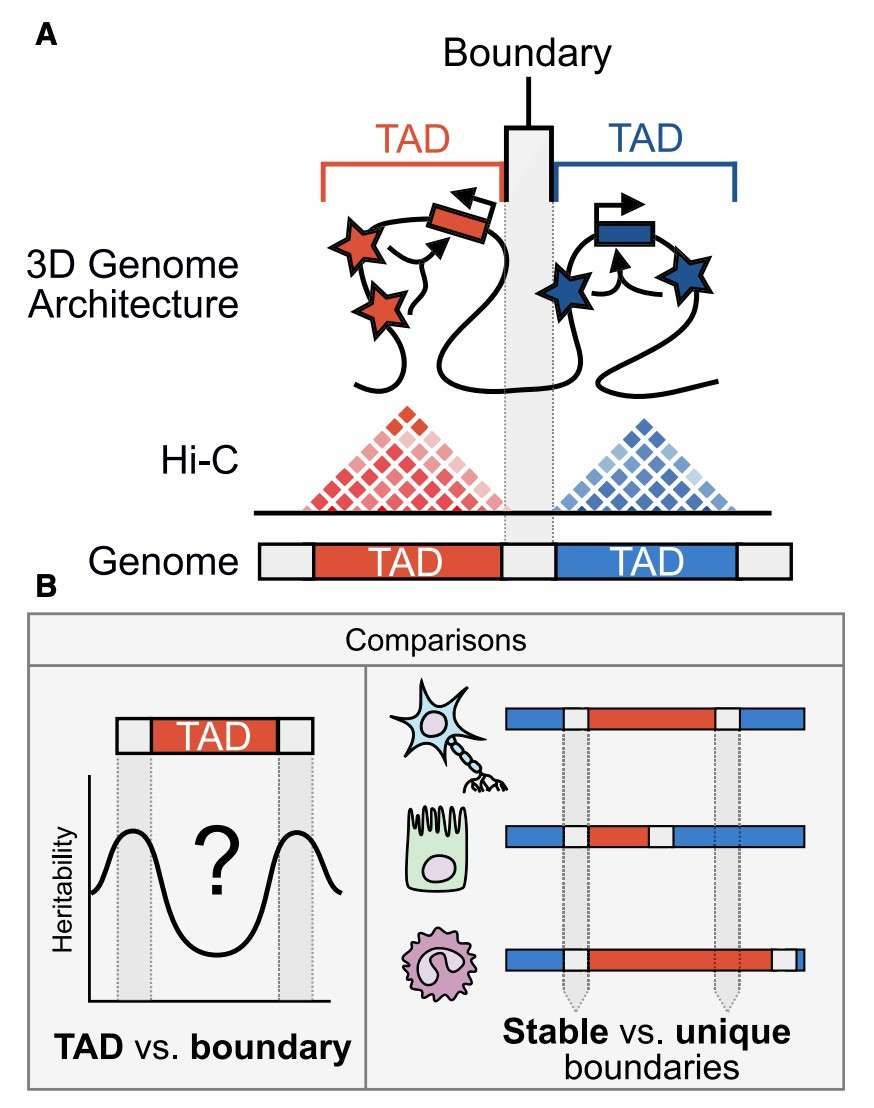 Schematic depiction of our analyses of 3D chromatin TAD-boundary stability and function (McArthur and John, 2021)
Schematic depiction of our analyses of 3D chromatin TAD-boundary stability and function (McArthur and John, 2021)
To further enhance the resolution of this technology, single-cell ATAC-Seq (scATAC-Seq) was developed. This technology can detect open regions of chromatin at the single-cell level and reveal the effect of cellular heterogeneity on chromatin remodeling. For example, in the context of embryonic development, scATAC-Seq data show that differences in the activity of NuRD complexes within the same class of stem cells may determine their fate to differentiate into neuronal or muscle cells. The integration of scATAC-Seq with single-cell transcriptome data facilitates the resolution of the dynamic correlation between chromatin opening state and gene expression, thereby providing novel perspectives for stem cell differentiation studies.
Tracking the dynamics of epigenetic modifications: from DNA methylation to RNA modification
DNA methylation (e.g., 5mC) and hydroxymethylation are significant markers of chromatin silencing and activation. Whole genome sulfite sequencing (WGBS), regarded as the "gold standard" for methylation analysis, can comprehensively cover all CpG sites in the genome and accurately map methylation. In the context of cancer, for instance, WGBS identifies aberrant hypermethylation of oncogene promoters that predate mutations, thereby providing markers for early diagnosis.
In the context of hydroxymethylation studies, 5hmC Selective Chemical Labeling Sequencing (5hmC-Seal Seq) has emerged as a pivotal technique, owing to its ability to specifically label the 5hmC locus. This approach has led to significant findings, including the observation that 5hmC levels in the cerebrospinal fluid of Alzheimer's disease patients were markedly reduced, exhibiting a negative correlation with the degree of Tau protein phosphorylation. These findings suggest that the restoration of 5hmC modification may emerge as a novel therapeutic strategy for treating Alzheimer's disease and related conditions.
Recent studies have revealed the involvement of RNA modifications (e.g., m6A, m5C) in the regulation of chromatin. MeRIP-Seq, a technique that enriches m6A-modified RNAs using antibodies, in conjunction with transcriptomic data, has elucidated the manner in which RNA epimodifications influence the recruitment of chromatin remodeling complexes. For instance, m6A-modified long non-coding RNAs (lncRNAs) have been observed to direct Polycomb complexes to specific genomic regions, resulting in the silencing of developmentally relevant genes. Conversely, ONT Direct RNA Sequencing has the capacity to directly read the full-length sequence of RNAs without the need for fragmentation, while detecting modifications such as m6A, thereby unveiling the cross-layer regulation of RNA epitope modification and chromatin structure. Through the integration of DNA methylation (WGBS), RNA modification (MeRIP-Seq), and chromatin accessibility (ATAC-Seq) data, scientists are able to construct a comprehensive regulatory chain, spanning from epigenetic modification to gene expression.
Real-time dynamic monitoring and precise intervention
The activity of chromatin remodeling complexes is highly dynamic. Live cell imaging techniques (e.g., CRISPR LiveFISH) allow real-time observation of the aggregation and dissociation of Polycomb complexes by fluorescently labeling specific DNA sites. Recent studies have revealed that Polycomb maintains the "memory" of gene silencing by forming dynamic cohesions through liquid-liquid phase separation.
In clinical translation, targeted DNA methylation analysis (EpiTYPER) provides an efficient tool for early cancer screening by quantifying the methylation level of specific CpG sites through mass spectrometry. For example, hypermethylation of the APC gene promoter in the blood of colorectal cancer patients can be detected at an early asymptomatic stage by EpiTYPER, with a 3-fold increase in sensitivity over traditional markers. For micro samples such as puncture biopsies or circulating tumor DNA (ctDNA), EM-seq technology optimizes the sulfite treatment process to complete genome-wide methylation analysis in only 100 cells, providing a solution for studies with scarce clinical samples.
Service you may intersted in
Learn More:
Clinical Targeting of Remodeling Complex Dysregulation
Mutations in chromatin remodeling complexes have been demonstrated to be closely associated with cancer and neurodegenerative diseases. In order to understand these associations, it is necessary to possess not only knowledge of molecular biology, but also precise epigenetic analysis tools. These tools act as "molecular diagnostics," which assist scientists in sifting through massive amounts of data to identify therapeutic targets.
Mutations in the BAF complex: cancer's "chromatin lock"
The BAF complex, a pivotal member of the SWI/SNF family, has been found to harbor mutations in its subunit ARID1A in up to 20% of ovarian and gastric cancers. In the absence of ARID1A, the BAF complex functions akin to a locksmith who has misplaced the key, rendering it incapable of opening the chromatin to facilitate the expression of oncogenes (e.g., p53). In this context, the emergence of EZH2 inhibitors, such as Tazemetostat, has marked a significant advancement in the treatment of follicular lymphoma. These inhibitors function by impeding the methyltransferase activity of the Polycomb complex and consequently reactivating silenced genes.
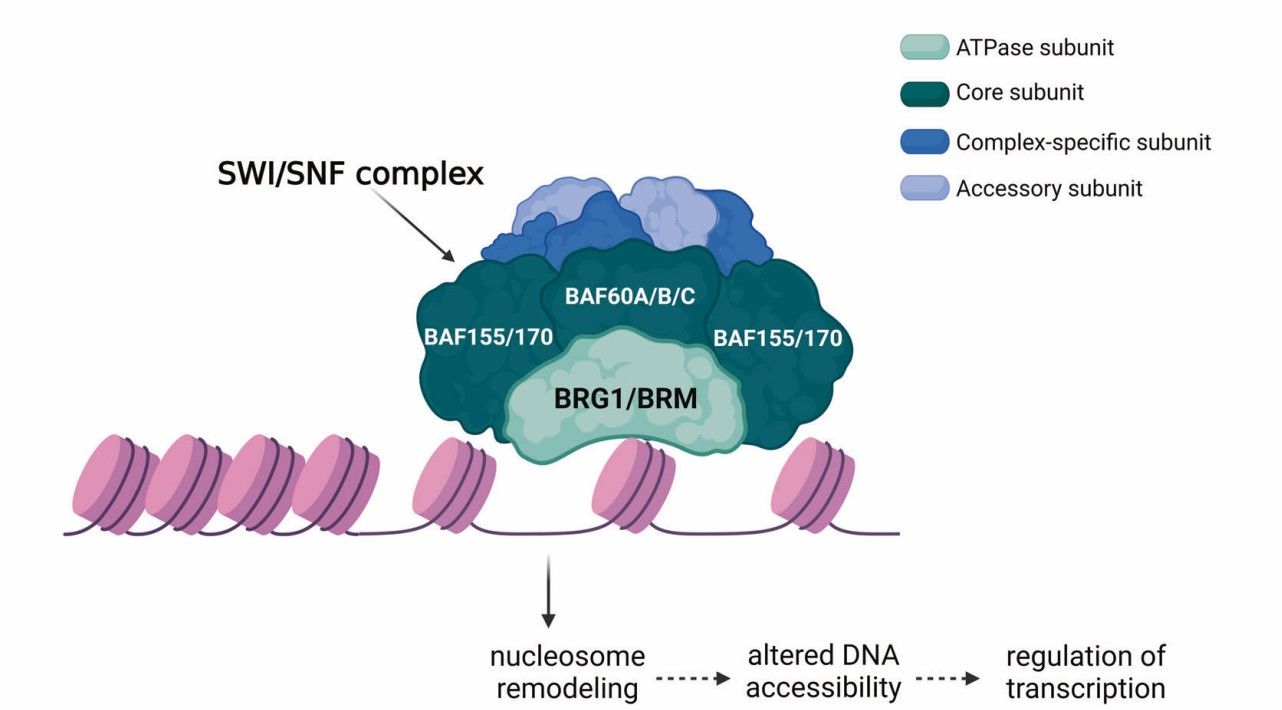 The SWI/SNF chromatin remodeling complex regulates gene expression (Navickas et al., 2023)
The SWI/SNF chromatin remodeling complex regulates gene expression (Navickas et al., 2023)
To identify suitable beneficiary populations, clinical laboratories frequently utilize targeted DNA methylation analysis (e.g., EpiTYPER). This technology employs mass spectrometry to quantify methylation levels at specific CpG sites. In the context of colorectal cancer, for instance, EpiTYPER can expeditiously discern hypermethylation of the APC gene promoter, an epigenetic silencing that frequently precedes gene mutations, serving as an early diagnostic marker.
Precision medicine: from mutation detection to targeted intervention
In the domain of neurodegenerative diseases, 5hmC-Seal Seq has emerged as a prominent research focus.5hmC, an intermediate product of DNA demethylation, is found in high concentrations in neurons. Utilizing this technique, scientists have observed that 5hmC levels are significantly reduced in the cerebrospinal fluid of individuals diagnosed with Alzheimer's disease. This reduction in 5hmC levels exhibits a negative correlation with the degree of phosphorylation of Tau protein. These findings suggest that the restoration of 5hmC modifications could serve as a novel therapeutic strategy to decelerate disease progression.
In the context of limited clinical sample sizes, such as puncture biopsies, microcellular whole genome methylation sequencing (EM-seq) has been shown to offer distinct advantages. This technology enhances the conventional sulfite processing step and facilitates the acquisition of reliable data from as few as 100 cells. A study on lung cancer demonstrated the efficacy of EM-seq in detecting specific methylation of the SOX17 gene in circulating tumor DNA. This epigenetic feature emerges earlier than conventional tumor markers, offering a novel approach for liquid biopsy.
Conclusion
Recent technological advancements, such as 3D genome mapping (Hi-C, scATAC-Seq) and dynamic tracking of epigenetic modifications (WGBS, 5hmC-Seal Seq), are contributing to the development of multi-dimensional and high-precision chromatin research methodologies. For instance, integrated multi-omics analysis (e.g., ATAC-Seq+RNA-Seq+methylation data) can elucidate the mechanisms by which chromatin remodeling complexes coordinate regulation at various epigenetic levels. Furthermore, artificial intelligence (AI)-driven Epigenomic Peak Calling (EPC) automatically identifies functional elements and predicts disease-associated variants from voluminous data sets.
In the future, with long-read and long-sequencing technologies resolving complex genomic regions, single-cell epigenomics revealing cellular heterogeneity, and liquid biopsy technologies (e.g., cfDNA methylation analysis) enabling non-invasive diagnosis, the field of chromatin remodeling research will be more closely integrated with clinical needs, providing the underlying technical support for precision medicine.
References
- Valdez, Benigno C et al. "Combination of a hypomethylating agent and inhibitors of PARP and HDAC traps PARP1 and DNMT1 to chromatin, acetylates DNA repair proteins, down-regulates NuRD and induces apoptosis in human leukemia and lymphoma cells." Oncotarget. 9,3 3908-3921. 17 Dec. 2017, doi:10.18632/oncotarget.23386
- Toto, Maria et al. "Regulation of ISWI chromatin remodelling activity." Chromosoma. 123,1-2 (2014): 91-102. doi:10.1007/s00412-013-0447-4
- Navickas, Sophie M et al. "The role of chromatin remodeler SMARCA4/BRG1 in brain cancers: a potential therapeutic target." Oncogene. 42,31 (2023): 2363-2373. doi:10.1038/s41388-023-02773-9
- McArthur, Evonne, and John A Capra. "Topologically associating domain boundaries that are stable across diverse cell types are evolutionarily constrained and enriched for heritability." American journal of human genetics. 108,2 (2021): 269-283. doi:10.1016/j.ajhg.2021.01.001
! For research purposes only, not intended for clinical
diagnosis, treatment, or individual health assessments.
 Suggested mechanisms of synergistic cytotoxicity (Valdez et al., 2017)
Suggested mechanisms of synergistic cytotoxicity (Valdez et al., 2017) Regulation of ISWI chromatin remodellers by histones and post-translational modifications (Toto et al., 2014)
Regulation of ISWI chromatin remodellers by histones and post-translational modifications (Toto et al., 2014) Schematic depiction of our analyses of 3D chromatin TAD-boundary stability and function (McArthur and John, 2021)
Schematic depiction of our analyses of 3D chromatin TAD-boundary stability and function (McArthur and John, 2021) The SWI/SNF chromatin remodeling complex regulates gene expression (Navickas et al., 2023)
The SWI/SNF chromatin remodeling complex regulates gene expression (Navickas et al., 2023)


