We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy

Single-cell Hi-C sequencing represents a transformative technology that has been developed for the purpose of investigating the three-dimensional organization of chromosomes in individual cells. The technique combines traditional Hi-C methods with the capability of single-cell sequencing, thereby enabling high-resolution analysis of genomic structure at the single-cell level. This method provides a novel means by which to study chromatin interactions in individual cells and consequently facilitates the construction of a precise three-dimensional genome structure specific to each cell.
The Underlying Principle of Single-Cell Hi-C
The principle of single-cell Hi-C technology is predicated on the Hi-C technology for resolving the three-dimensional spatial structure of chromosomes in a single cell. Initially, researchers extract chromosomes from single cells and treat the cells with chemical reagents such as formaldehyde, so that deoxyribonucleic acid (DNA) fragments with similar spatial distances on the chromosomes, as well as biological macromolecules such as proteins bound to them, undergo a cross-linking reaction and fix the three-dimensional structure of the chromosomes. Subsequently, restriction endonuclease is utilised to enzymatically cleave the cross-linked chromatin DNA into numerous fragments of varying sizes. In the subsequent step, ligase is employed to join the sticky ends of the enzymatically cleaved DNA fragments to form a circular DNA molecule. The next stage of the process involves the labeling of the DNA ring using markers such as biotin, and the purification of the labeled DNA using substances with specific binding ability, such as affinity factors, by methods such as affinity chromatography, to remove unconnected or non-specifically connected DNA fragments. Finally, high-throughput sequencing is performed on the purified DNA to obtain a large amount of DNA sequence data. The resulting data were then analysed using bioinformatics algorithms and software, which enabled the comparison of the sequencing data with the reference genome. This comparison was used to study the linkage relationship between DNA fragments, and thereby infer the three-dimensional spatial structure of chromosomes in a single cell. This structure includes features such as chromatin loops and topologically related structural domains. The use of single-cell Hi-C has been shown to provide researchers with a detailed understanding of the organisation of chromosomes in space and their variability in different cells.
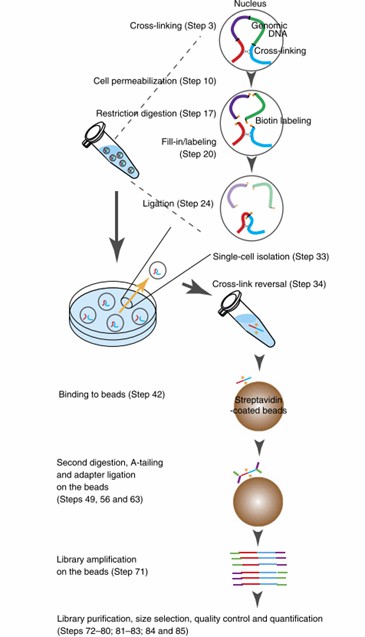 Overview of the single-cell Hi-C protocol (Nagano et al., 2015)
Overview of the single-cell Hi-C protocol (Nagano et al., 2015)
Single-cell Hi-C technology finds broad applications across various research domains, especially in genomics, cancer biology, and developmental biology, by providing detailed insights into chromatin structure at the cellular level.
Genomic Applications
In genomics, single-cell Hi-C is instrumental in detecting chromosomal structural variations. Through the analysis of single cells, researchers have uncovered subtle genomic alterations such as chromosomal rearrangements, copy number variations (CNVs), and other structural anomalies. This ability to capture a three-dimensional view of the genome's architecture provides a deeper understanding of spatial variations in chromatin organization across cell types. Studies have shown that chromosomal topological domains exhibit variability across different cell types and stages of the cell cycle, offering fresh perspectives on genomic function and gene regulation.
Developmental Biology
In developmental biology, single-cell Hi-C plays a crucial role in examining the alterations in chromosomal structure during cellular differentiation and embryonic development. Research has revealed significant transformations in chromatin structure during development, closely tied to gene expression regulation. Single-cell Hi-C allows for tracking these structural changes across various developmental stages, helping scientists to gain a comprehensive understanding of how genes are spatially organized and regulated during these processes. This technology serves as a powerful tool for studying how cellular identities are established and how they contribute to complex developmental phenomena.
Cancer Research
Single-cell Hi-C is especially promising in cancer research, as it allows for the exploration of tumor heterogeneity and chromosomal rearrangements within cancer cells. Cancer cells frequently experience chromosomal changes that alter gene expression, promote tumor growth, and contribute to metastasis and drug resistance. Single-cell Hi-C enables detailed analysis of chromatin organization within individual tumor cells, identifying aberrant chromatin interactions. By analyzing various tumor cell types, researchers can detect unique chromosomal translocations and structural rearrangements, which are essential for understanding cancer progression and developing personalized treatment approaches.
Service you may intersted in
The application of single-cell Hi-C has resulted in substantial breakthroughs across multiple research areas, particularly genomics, cancer biology, and developmental studies. These case studies highlight the transformative potential of this technology.
In 2020, Vijay Ramani and colleagues utilized single-cell Hi-C to investigate mouse and human cell lines. By employing a 96-well plate for dual barcoding, they labeled and mapped chromatin interactions in thousands of single cells. The results showed that single-cell Hi-C could distinguish different cell types based on their chromatin conformations, unveiling heterogeneity in chromosomal structure across cells. This work demonstrated the power of single-cell Hi-C in understanding genomic variation and chromatin organization, providing valuable insights into cell cycle dynamics, cancer biology, and cellular development.
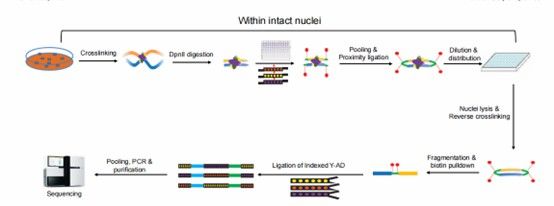 The workflow of sci-Hi-C (Ramani et al., 2020)
The workflow of sci-Hi-C (Ramani et al., 2020)
A significant investigation was conducted by Takashi Nagano and colleagues, who analyzed the chromatin structure of mouse T-helper cells using the single-cell Hi-C technique. This technique was used to examine the chromosomal organization and spatial interactions within individual cells. It is hoped that, by conducting this research, a more comprehensive insight will be gained into the complex interplay between genetic composition and cellular function. The findings of this research revealed that individual chromosomes maintain topological domain organization at the megabase scale, but structural variations were observed at larger scales. This offers valuable insights into the dynamics of chromosome organization and the potential implications for cellular processes and diseases. This research has provided new tools for exploring chromosomal heterogeneity and the three-dimensional organization of the genome, helping to deepen our understanding of cell-specific gene expression and disease mechanisms.
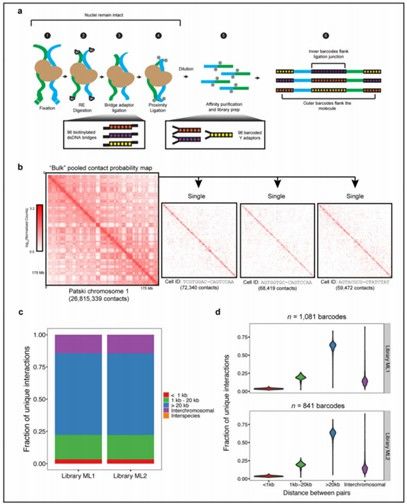 Single-cell combinatorial indexed Hi-C integrates the in situ Hi-C protocol with cellular indexing to generate signal-rich bulk Hi-C maps that can be decomposed into single-cell Hi-C maps (Ramani et al., 2017)
Single-cell combinatorial indexed Hi-C integrates the in situ Hi-C protocol with cellular indexing to generate signal-rich bulk Hi-C maps that can be decomposed into single-cell Hi-C maps (Ramani et al., 2017)
Single-cell Hi-C sequencing offers numerous advantages but also presents several challenges that need to be addressed for its broader application.
Advantages of Single-Cell Hi-C
The primary advantage of single-cell Hi-C lies in its ability to analyze the three-dimensional structure of chromosomes at the individual cell level, providing in-depth insights into the spatial organization of the genome. Unlike traditional Hi-C, which averages chromatin interactions across populations of cells, single-cell Hi-C can capture the heterogeneity between cells. This allows for the study of how different cell types, stages of the cell cycle, or disease states impact chromatin structure. Furthermore, the high resolution of single-cell Hi-C enables the detection of subtle structural variations such as gene copy number changes and chromosomal translocations. This makes single-cell Hi-C a powerful tool in genomics, cell biology, and disease research.
Technical Challenges
Despite its many advantages, single-cell Hi-C presents several technical challenges. The first major challenge is the effective isolation of single cells while maintaining the integrity of their chromatin. While advancements in microfluidics and cell separation technologies have improved this process, isolating high-quality single cells remains a critical hurdle. Additionally, single-cell Hi-C generates vast amounts of complex data, and the efficient analysis of this data remains challenging. Most existing algorithms are optimized for population-level data, and new methods are needed to handle single-cell datasets with high accuracy and resolution.
The future of single-cell Hi-C sequencing focuses on improving the technology and expanding its applications.
Technological Advancements and Future Outlook
As sequencing technologies improve, the resolution of single-cell Hi-C will continue to enhance, allowing for even finer detection of chromosomal changes. Automation of experimental procedures will further streamline workflows, increasing efficiency and reducing costs. These technological advancements are set to elevate the role of single-cell Hi-C in studying chromosomal structure, gene expression regulation, and disease mechanisms.
Integration with Other Omics Data
The integration of single-cell Hi-C with other omics data, such as transcriptomics and epigenomics, will provide a more comprehensive understanding of the genome. By combining data from various sources, researchers can gain deeper insights into how chromatin structure affects gene expression and cellular functions. This cross-omics approach will generate novel perspectives on the dynamic behavior of cells and the intricate regulation of the genome.
The advent of biotechnology has precipitated the increased utilisation of single-cell Hi-C sequencing in biological research, offering novel insights. This technique facilitates the analysis of the spatial configuration of chromosomes at the single-cell level, thereby unveiling chromosomal heterogeneity among cells. It has significant applications in cancer research, developmental biology, and elucidating disease mechanisms. Notwithstanding the challenges pertaining to technical complexity and economic burden, single-cell Hi-C is poised to assume an increasingly prominent role in future biological applications, as the technology undergoes optimisation and evolves towards more sophisticated applications.
References
Terms & Conditions Privacy Policy Copyright © CD Genomics. All rights reserved.