Technology Introductions
Chromatin immunoprecipitation followed by sequencing (ChIP-seq) is a method used to study protein-DNA interactions. The technique combines chromatin immunoprecipitation (ChIP) with NGS to identify the site of DNA binding to the protein of interest, allowing precise localization of the binding site of the target protein on the whole genome. The DNA-protein binding complex is immobilized in live cells, then the complex is captured by immunological means of antigen-antibody specific binding with protein-specific antibodies, and then the proteins are eluted to obtain DNA fragments bound to the target protein, and the enriched DNA fragments are subjected to on-line sequencing. Compared to chip-based Chip-chip technology, ChIP-seq experiments are shorter and more efficient, and cover a broader genome. The reduced cost of sequencing makes ChIP-seq one of the tools for epistasis research.
Specific CHIP-Seq Approaches We Offer
Library construction
Regarding ChIP-seq library construction and sequencing, the main steps are as follows:
1. Formaldehyde cross-linking of cell lines to ligate target proteins to chromatin.
2. Isolating genomic DNA and breaking it into small fragments of a certain length by ultrasound/enzymatic digestion.
3. Addition of antibodies specific to the target protein to form an immunoprecipitated immunobinding complex of antibody and target protein.
4. De-crosslink, purify DNA and build library for sequencing.
Analysis process
For ChIP-seq data, the raw data were firstly evaluated for quality using fastqc software, then the raw data were compared to the reference genome using BWA or bowtie software, and then peak calling was performed for each sample separately using MACS, followed by genomic position annotation, motif analysis for the peaks region, and target If there are multiple experimental groups of samples, we can mine the peaks with differences between the samples for specificity analysis.
Service Advantages
- Map genome-wide histone protein modifications using CD Genomics' comprehensive chromatin immunoprecipitation sequencing (ChIP-Seq) platform. Optimized buffers and protocols minimize ChIP background and increase the sensitivity and specificity of ChIP reactions.
- Highly efficient enrichment of target DNA with an enrichment ratio of > 500 for positive to negative controls.
- High sensitivity and flexibility allows for non-barcoded (single-duplex) and barcoded (multiplex) DNA library preparation.
Our Capabilities
- Sequencing and library preparation assurance
Library quality control by bioanalyzer, KAPA quantification and Q30 sequencing scores > 75%
- Histone modifications included
Use any H2, H3, H4 histone modification from our catalog at no additional cost
ChIP-level antibody validation step demonstrates specificity, affinity and reproducibility. affinity and reproducibility
- Flexible sample requirements
ChIPed DNA with histone modifying antibodies is acceptable
- Post-completion technical support
Free, high-quality support to assist with data analysis at no additional cost
Workflow
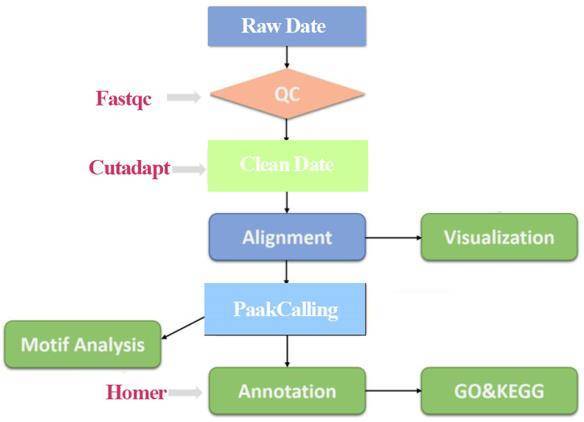
Sample Requirements
Cells required for ChIP-seq reaction should be greater than 10million/tube.
Select fresh tissues with mass greater than 100mg/serving, snap frozen in liquid nitrogen and stored at -80℃. It is recommended that samples be prepared in 2~3 portions.
The mass is more than 5g/sample. It is recommended to rinse the sample with water to remove the dirt, blot the water on paper towel and then snap freeze it in liquid nitrogen and store it at -80℃. It is recommended to prepare 2~3 samples.
Service Process

Deliverables
- Related experimental results raw data
- Experimental report
- Data analysis
- Image and result analysis
- Bioinformatics analysis results
- Details in CHIP-Seq for your writing
Our Features
- We have accumulated many years of essence in the field of CHIP-Seq service, helping customers to speed up project development and improve the overall success rate of the project.
- At CD Genomics, simply let us know your specific project needs. We will suggest the best strategy for you.
Why Choose Us?
CD Genomics is a company that provides professional and comprehensive CHIP-Seq services. We have years of experience to meet your specific project needs in using epigenomics research to add value to your research projects. CD Genomics can provide you with personalized solutions to help you thrive every step of the way around your interest in your workflow. If you would like to know more about this service, please feel free to contact us.


