Technology Introduction
ATAC-seq (Assay for Transposase-Accessible Chromatin with high throughput sequencing), which inserts sequence junctions into chromatin open regions by hyperactivating Tn5 transposase, and then analyzes DNA sequencing data to infer the accessibility of chromatin regions, transcription factor binding sites and nucleosome locations.
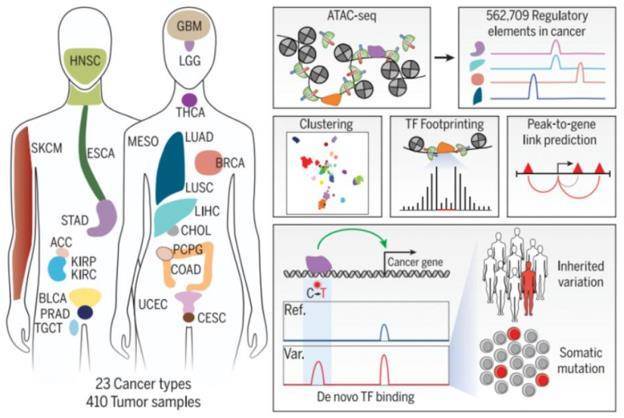 Figure 1. Genomic distribution characteristics and number of cancer types, sample sizes and peaks. (Corces M R, et al.2018)
Figure 1. Genomic distribution characteristics and number of cancer types, sample sizes and peaks. (Corces M R, et al.2018)
Specific ATAC-Seq Approaches We Offer
ATAC-seq library construction
- Enrichment of cells for lysis and extraction of purified nuclei.
- Performing Tn5 enzyme transposition reaction to fragment the chromatin open region.
- Purify the fragmented DNA.
- Amplify the purified product.
- Sort the amplified DNA length and remove the non-target fragments.
Analysis process
For ATAC-seq data, CD Genomics first uses fastqc software to assess the quality of the raw data; then uses bowtie2 software to match the raw data to the reference genome; then uses MACS2 to perform peak calling for each sample separately, followed by genomic position annotation, motif analysis, and GO and KEGG enrichment analysis for the peaks region. If there are multiple experimental groups of samples, the peaks with differences among the samples can be mined for specificity analysis, etc.
Services Advantages
- Compared with conventional methods, ATAC-seq results are highly consistent and also have a high degree of agreement with ChIP-seq based on histone modification markers.
- The reproducibility of ATAC-seq is stronger and simpler to perform than MNase-seq and DNase-seq.
- Less cell or tissue is required and comes out with a more beautiful signal.
- ATAC-seq is currently the preferred technical method for studying chromatin openness.
Our Capabilities
- CD Genomics can develop its own tissue protection solution to guarantee cell activity for long-distance transport.
- CD Genomics can develop its own ATAC-seq cell protection solution and ATAC-seq tissue protection solution, guaranteeing cell activity for 48 hours long-distance transport.
- Strict control of mitochondrial contamination at less than 5%.
- Personalized bioinformatics analysis, which can provide a variety of personalized implementation solutions.
Sample Requirements
- Sample type: fresh cells, quantity 1 x 103- 105
- Fresh tissue, mass of 50 mg/ serving
- Sequencing mode: illumina Hiseq PE150
- Recommended data volume: ≥10Gb/ sample
Service Process

Deliverables
1. Related experimental results raw data
2. Experimental report
3. Data analysis
4. Image and result analysis
5. Bioinformatics analysis results
6. Details in ATAC-Seq for your writing
Our Features
- CD Genomics has a group of technical experts specializing in ATAC-seq services. Our extensive research and technical experience can help our customers solve their ATAC-seq technical problems.
- CD Genomics has a proven, advanced ATAC-seq technology platform that is constantly being improved to ensure that we can provide accurate, sensitive, and rapid analytical results for our customers.
Why Choose Us?
CD Genomics is a professional ATAC-seq technology provider supporting a wide range of ATAC-seq services. We are able to perform individual tests at the most precise level, as well as design and complete a suite of services as a solution to the subject project. We have accumulated years of quintessence in the field of ATAC-seq service, helping our clients accelerate drug discovery and development and improve the overall success rate of their projects. If you would like to know more about this service, please feel free to contact us.
Reference
- Corces M R, Granja J M, Shams S, et al. The chromatin accessibility landscape of primary human cancers. Science, 2018, 362(6413):420-420.

 Figure 1. Genomic distribution characteristics and number of cancer types, sample sizes and peaks. (Corces M R, et al.2018)
Figure 1. Genomic distribution characteristics and number of cancer types, sample sizes and peaks. (Corces M R, et al.2018)