
Navigation
- Home
- Services
- Microbial Diversity Analysis – 16S/18S/ITS Sequencing
- Metagenomics
- Microbial Whole Genome Sequencing
- Microbial Identification
- Microbial Characterization
- Microbial Functional Gene Analysis
- Microbial Epigenomics
- Antibiotic Resistance Genes (ARGs) Analysis Solution
- Microbial PacBio SMRT Sequencing
- Microbial Nanopore Sequencing
- Microbial Transcriptomics
- Microbial Bioinformatics
- Other
- Microbial Metabolomics Analysis Service
- Solutions
- Microecology and Human/Animal Health
- Environmental Microbiology Solutions
- Agricultural Microbiology Solutions
- Pharmaceutical Microbiology Solutions
- Industrial Microbiology Solutions
- Microecology and Cancer Research Solutions
- Microecology and Biofilms
- Pathogen Sequencing Solutions
- Environmental DNA (eDNA) Analysis Solution
- Products
- Resource
- Company
- Sample Submission Guidelines


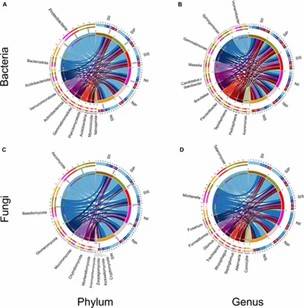 Fig 1. Relative abundance of the peanut rhizosphere (IP), sorghum rhizosphere (IS), and interspecific interaction zone (II) microbial communities at the phylum and genus levels under different soil conditions.
Fig 1. Relative abundance of the peanut rhizosphere (IP), sorghum rhizosphere (IS), and interspecific interaction zone (II) microbial communities at the phylum and genus levels under different soil conditions.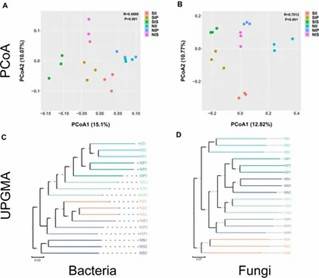 Fig 2. Principal coordinate analysis and unweighted pair group method with arithmetic mean analysis.
Fig 2. Principal coordinate analysis and unweighted pair group method with arithmetic mean analysis.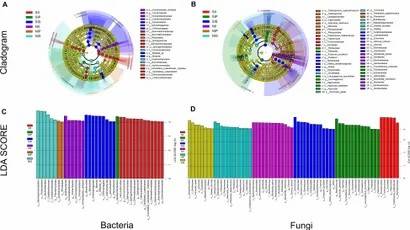 Fig 3. Linear discriminant analysis effect size.
Fig 3. Linear discriminant analysis effect size.








