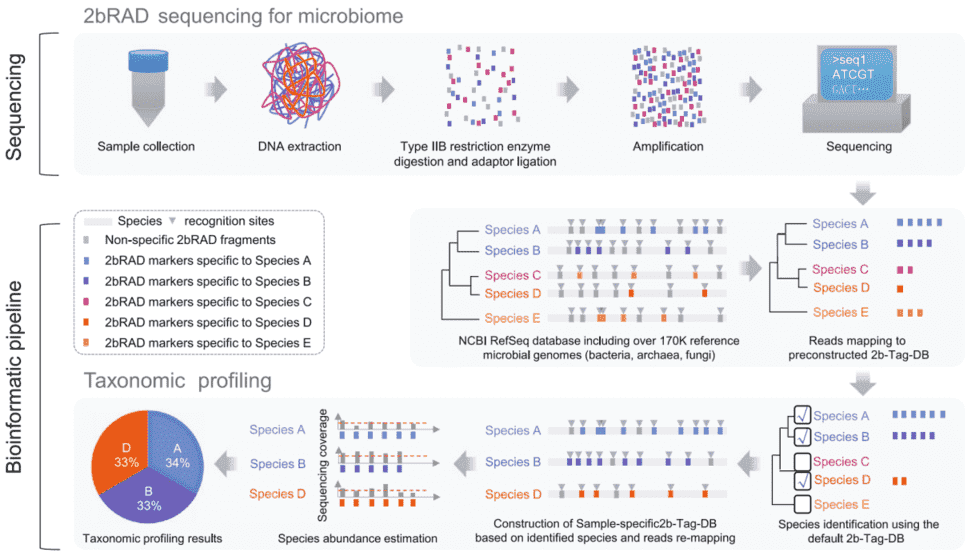
A new microbiome sequencing technology—2bRAD-M, originated from 2bRAD, a streamlined restriction site-associated DNA (RAD) genotyping (Nature Methods, Wang et al. 2012), which digests microbial genomes with type IIB restriction endonucleases after obtaining the unique tag (unique tag), and then qualitative and relative quantitative analysis of microorganisms, this technology has obvious technical characteristics and advantages compared with traditional microbiome sequencing technology.
Technical Features and Advantages
- Simultaneous detection of bacteria, fungi and archaea in a single sequencing
- Suitable for difficult samples: low biomass (pg level DNA), severely degraded samples (e.g. FFPE), high host contamination samples, etc.
- High sensitivity: high detection capacity even for low abundance microbial species
- Species level resolution for species detection

Tell Us About Your Project
We are dedicated to providing outstanding customer service and being reachable at all times.
 Request a Quote
Request a Quote
Technical Principle
The genome is cleaved by IIB type restriction endonuclease, which can produce iso-length tags (e.g., 32bp equal length tag using BcgI enzyme), and the IIB tag is amplified and sequenced, and then the microbial community structure (including bacteria, fungi, and archaea) is analyzed by a "two-step" method.
In the first step, qualitative analysis is performed through a pre-established database (2b-Tag-DB) containing the unique tag of each microorganism, that is, to screen all microbial species that can detect unqiue tags.
The second step is to establish 2b-Tag-DB for qualitative microorganisms and perform relative quantitative analysis, that is, to further screen and estimate the abundance of the microbial species obtained in the previous step according to the unique tag distribution. This strategy can maximize the detection of microorganisms with high accuracy and sensitivity, and can detect bacteria, fungi and archaea simultaneously.
2bRAD-M FAQ
For microbiome sequencing, must the entire genome be sequenced to achieve species-level resolution?
Not really. 2bRAD-M technology simplifies the overall genome to achieve efficient cost control and achieve the resolution of microbial species level identification. Although 2bRAD-M is a simplified metagenomic sequencing technology that only obtains about 1% of genome information, this 1% of information is representative of the entire genome and also provides accurate microbial community structure.
For samples with high host contamination, metagenomics generally obtain a lot of useless data. Is it the same for 2bRAD-M?
2bRAD-M technology can overcome high host contamination for three reasons: (1) 2bRAD-M greatly reduces the overall sequencing cost of samples by sequencing a representative 1% of the human genome and microbial genome, and greatly increases the sequencing depth of each individual organism. (2) Due to the high imbalance of restriction sites in microbial genomes and human genomes, microbial genomes can generate more 2bRAD tags than human genomes. For example, if the ratio of microbial DNA standard (Mock-CAS) to human DNA is 1:99, in the actual sequencing results, the ratio of sequencing reads between the two will become 3:97.
(3) The 2bRAD signature in the human genome is completely different from the 2bRAD signature in the microbial genome. Therefore, 2bRAD-M technology can effectively resist host DNA interference and greatly reduce the false positive rate. 2bRAD-M technology can handle up to 99% host-contaminated samples (below).
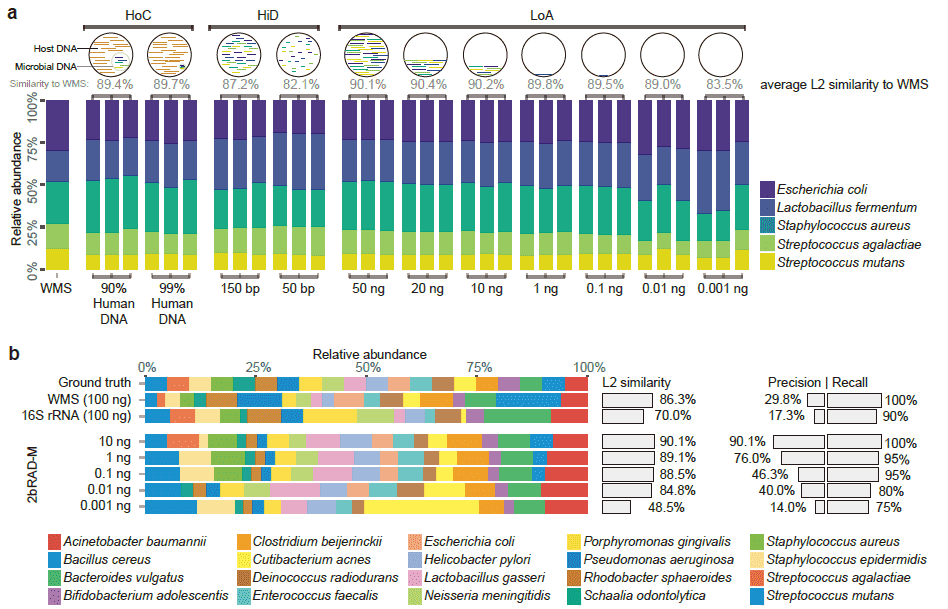
Is there a way to detect microorganisms in sample using 2bRAD-M if the biomass/nucleic acid amount is very low?
It is possible. It has been proven that 2bRAD-M technology can effectively handle low-biomass microbial samples, and can deal with samples with a total DNA of only 1pg, and has been successfully applied to the detection of low-biomass samples such as skin, indoor carpets, and car mats. Taking skin samples as an example, at the genus level, the microbial classification results of 2bRAD-M and 16S were highly consistent, and Staphylococcus was found to be the dominant microorganism; However, at the species level, 2bRAD-M can further find that the most abundant Staphylococcus is Staphylococcus epidermidis, and there is no significant difference among other species of this genus, which cannot be done by 16S. At the same time, 2bRAD-M can also detect fungi, which can reveal new bacterial-fungal interactions in low-biomass microbial communities at the species level.
Reference
- Sun, Z., Huang, S., Zhu, P. et al. Species-resolved sequencing of low-biomass or degraded microbiomes using 2bRAD-M. Genome Biol 23, 36 (2022).