Bacteria are prokaryotic life forms, typically unicellular, with a relatively simplistic cellular structure that typically exists on a micrometer scale. The structural architecture of a bacterial cell encompasses a cytoplasm, cell wall, and cell membrane, while some bacteria also exhibit appendages such as flagella or pili. Their genetic material, comprised typically of a singular chromosomal ring, resides within the cytoplasm, typically unattached to a nucleus. The primary energy acquisition strategy for bacteria hinges on the absorption and decomposition of organic materials, although there are certain bacterial species that harness light energy through photosynthesis. Bacteria pervade a wide expanse of environments in nature, including but not limited to soil, water, air, and both the exterior and interior of biological organisms, thus signifying their ecological importance and biological relevance. While certain bacteria play beneficial roles for humans and other organisms, serving integral roles in biogeochemical cycles and ecological balance, some bacterial species are pathogenic, capable of inducing diseases.

Fungi belong to the Eukaryotes, and compared to bacteria, their cellular structure is complex, typically multi-cellular, though some are unicellular organisms. The cellular structure of fungi comprises distinct components such as nucleus, cytoplasm, cell wall, and cell membrane. Their genetic material resides in the cell nucleus in the form of linear chromosomes. Fungi primarily obtain energy through the absorption of organic matter, typically decomposing organic materials for nutrient acquisition. Fungi are extensively distributed across all types of environments and ecosystems, such as soil, bodies of water, plant surfaces and internals, and within animal bodies. They play a significant role in ecosystem material cycling and energy flow. Some fungi are pathogenic and can cause diseases in plants and animals. Nevertheless, a large number of fungi benefit humans and other organisms, examples of which include edible mushrooms and brewing yeast.
Table 1. Differences Between Bacteria and Fungi
|
Bacterial |
Fungi |
| Definition |
Single-celled, prokaryotic organisms, relatively simple cell structure |
Multicellular, eukaryotic organisms, complex cell structure |
| Size |
0.5-5 μm |
2-10 μm |
| Characteristics |
Absence of organelles
Absence of organelles
Cell wall composed of peptidoglycan |
Presence of organelles
Presence of organelles
Cell wall composed of chitin |
| Morphology |
Three distinct shapes: spherical (cocci), spiral (spirilla), and rod-shaped (bacilli) |
Diverse shapes, mostly observed in filamentous structures called hyphae |
| Producer/Decomposer |
Can be both producers (chemoautotrophs, photoautotrophs) and decomposers (soil bacteria) |
Typically decomposers |
| Acidity/Alkalinity |
Thrive best in neutral environment with pH 6.5-7 |
Prefer slightly acidic environment with pH 4-6 |
| Nutritional Mode |
Autotrophic or heterotrophic |
Heterotrophic, feeding on dead and decaying matter |
| Reproductive Mode |
Asexual (binary fission) |
Both sexual and asexual |
| Motility |
Movement facilitated by flagella |
Non-motile |
| Genetic Material |
Located in nucleoid region within cytoplasm |
Located within nucleus |
| Nutrition |
Autotrophic or heterotrophic |
Mostly heterotrophic, feeding on dead and decaying matter |
| Energy Source |
Obtained from inorganic or organic compounds (e.g., sugars, proteins, or lipids) |
Obtained from pre-existing organic matter |
| Respiration |
Aerobic and anaerobic respiration |
Mostly ethanol fermentation or anaerobic respiration |
| Diseases Caused |
Leprosy, cholera, tuberculosis, tetanus |
Ringworm, athlete's foot, allergic bronchopulmonary aspergillosis |
| Examples |
Escherichia coli, Staphylococcus aureus, Salmonella typhi, Lactobacillus spp. |
Saccharomyces cerevisiae (brewer's yeast), Aspergillus niger, Agaricus bisporus |
Traditionally, the differentiation between bacteria and fungi primarily relies on their cellular morphology and colony configuration. However, with advances in sequencing technology, discrimination between bacteria and fungi can also occur via differences in their nuclear structures and genetic material. The following content elucidates this distinction of bacteria and fungi from the perspectives of colony morphology and sequencing methodologies. Sequencing methodologies involving Internal Transcribed Spacers (ITS), as well as 16S rRNA - both underpinned by Next-Generation Sequencing (NGS) - proffer non-cultivation approaches in the identification of bacteria and fungi. These robust methods can facilitate the detection of organisms potentially overlooked by other techniques.
What Is Bacterial Colonies
The bacterial colonies represent a copious population of bacterial cells proliferated from a single bacterium, residing compactly upon a nourishing medium. These microbial populations within the colony are genetically analogous, rendering these colonies akin to clones. A multitude of bacterial colonies exhibit a spherical or irregular morphology, while others mimic filamentous or mycelial forms akin to actinomycetes. Innumerable colonies portray extreme minuteness, with diameters measuring less than 1 millimeter, earning them the epithet "punctiform," or pinpoint. To facilitate an observation of such minute protrusions, utilization of a microscope is recommended. The coloration of colonies often varies, species-dependent, with hues ranging from stark white to pale yellow apparitions, with shades of violet and purple interspersed.
Bacterial colonies may exhibit a myriad of surface textures - they might be glossy and smooth-surfaced, coarse, matte finished, or exhibit unevenness reminiscent of wrinkling. The tactile property of these colonies can differ drastically from a buttery consistency, to a sticky texture that clings tenaciously to the loop and proves difficult to dislodge, to a brittle or fragile nature, which could indicate desiccation or fragmentation, and finally, to a viscous, mucus-like texture.
What Is Fungal Colonies
Fungi, encompassing yeasts, filamentous fungi, and mushrooms, constitute a collection of eukaryotic species. Thriving best under moist, warm conditions, they can be classified based on their morphological and molecular attributes. Cultivating fungi on solid medium, such as Potato Dextrose Agar (PDA), allows for clear morphological characterization, making PDA an often-used tool for fungal culture in laboratories. During stable growth on such medium, fungi form colonies, each with distinctive morphology. Fungal colonies exhibit different morphologies when compared to bacterial colonies. They appear as powdery or hairy textured colonies. Fungal hyphae penetrate the solid medium forming pseudoroots or filamentous colonies as opposed to appearing as bacterial colonies which might be represented as tiny oil dots. The pigmentation of hyphae and spores also vastly varies across different species of fungus.
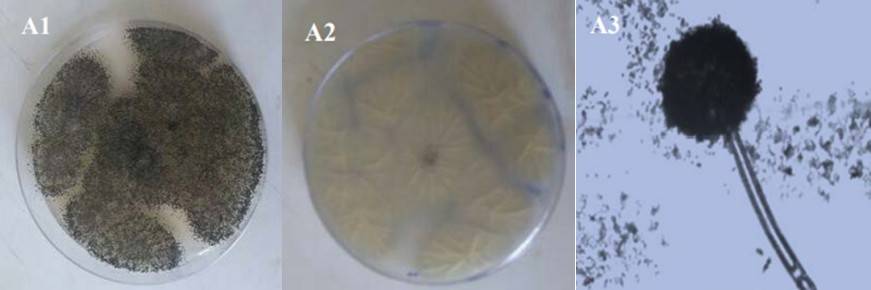 Figure 1. Aspergillums niger colony features on PDA (A1 Top, A2 Reverse) and conidia (A3). (Alsohaili et al., 2018)
Figure 1. Aspergillums niger colony features on PDA (A1 Top, A2 Reverse) and conidia (A3). (Alsohaili et al., 2018)
Both bacterial and fungal colonies can proliferate on a solid nutrient agar medium. Their formation is representative of organisms displaying a communal trait. The coloration of these colonies depends on the species of microorganisms present, which can be utilized in microbial detection and diagnosis. However, their morphologies and growth patterns vary significantly, providing distinctive visual cues for their differentiation and identification.
Differences Between Bacterial and Fungal Colonies
In the classification of bacteria and fungi, morphological features hold significant relevance. Colony morphology serves as a useful tool widely employed by researchers in their endeavours to classify and define various bacterial and fungal colonies. In these investigative works, the specific characteristics of individual bacterial and fungal colonies are meticulously studied and utilized. Compared to fungi, bacteria tend to proliferate easily in nutrient-rich mediums. Colonies of unique phenotypes are formed by a diverse variety of bacteria and fungi. These colonies exhibit varying sizes, shapes, textures, colours, and margins. Bacteria and fungi should be cultured on agar mediums placed within petri dishes, with all necessary nutrients and conditions provided to examine the colony morphology.
Upon these agar mediums, bacteria manifest as microscopically tiny oily spot formations. Conversely, fungi manifest as a powdery mat spreading across the entire surface of the agar plate. A primary distinction between bacterial and fungal colonies arises from their origins. A recognizable cluster of bacterial cells emerging from a single bacterial cell constitutes a bacterial colony, while a visible fungal mass appearing from a single spore or mycelium fragment constitutes a fungal colony.
Table 2. Differences Between Bacterial and Fungal Colonies
|
Bacterial colonies |
Fungal colonies |
| Definition |
Bacterial colonies arise from the proliferation of individual bacterial cells on the surface of a nutrient-rich medium |
Fungal colonies emerge through the reproduction of individual fungal spores or hyphae on the surface of a culture medium containing appropriate nutrients and humidity |
| Number of cells |
Consist of unicellular cells |
Consist of unicellular or multicellular |
| Composition |
Consist of a mass of bacterial cells arising from a single bacterium's fragmentation |
Consist of fungal hyphae made up of a single spore |
| Colony size |
Tiny |
Huge |
| Appearance |
A smooth or rough texture |
Have a hairy look |
| Margin |
Have a fixed margin |
A filamentous margin |
| Texture |
Damp and glossy |
Powder-like |
| Shape |
Spherical or irregular |
Filamentous or rhizoid |
| pH compatibility |
Develop within ph 5-9 (optimum 7) |
Within ph 5-6 |
Colonies are identifiable as observable aggregates of microorganisms. Each colony has a parent cell, and the cells within the population are also genetically similar. Bacteria and fungi appear as colonies on solid mediums. On the agar surface, bacterial colonies appear as tiny creamy spots whereas fungal colonies form molds on the same surface. This constitutes the principal distinction between bacterial colonies and fungal colonies. Colony morphology contributes to the identification and differentiation of bacteria and fungi.
Differences Between Bacteria and Fungi in Sequencing
Microbial research was initially heavily dependent on morphological examination, a method that opened the doors to the diverse world of microorganisms and provided a foundation for rudimentary classification. While the process was effortful, its insights were inherently limited. With the evolution and refinement of sequencing technologies, microbiology has seen rapid advancements within a relatively short span of time. Techniques encompassing targeted sequencing, metagenomics, transcriptomics, and epigenomics have become integral to this exploration. These methods not only bolster efficiency but also significantly enhance accuracy. As our understanding of microbes deepen, research involving microbial metabolic pathways, gene utility, and microbial modifications have become increasingly more accessible.
- Differences between bacterial and fungal genomes
The genomic size of bacteria commonly falls in the range of 0.16-13Mb—typically around the 5Mb mark—distinguished by its structure consisting of a single circularized, closed double-stranded DNA, possessing no introns. Comparatively, bacteria maintain a low Guanine-Cytosine (GC) content. In contrast, the fungal genome size is generally distributed between 10Mb and 150Mb—most commonly approximately 50Mb. The structure of fungal genomic DNA extends linearly and, in most cases, presents a higher content of GC. The genomic complexity of fungi typically surpasses that of bacteria, encompassing a larger contingent of genes, non-coding regions, as well as an expanded repertoire of repeated sequences and gene families. These sequences play significant roles in gene replication and regulation, conversely seen less frequently in bacterial genomes.
- Difference between bacterial and fungal sequencing identification method
Distinct techniques are leveraged for the sequencing identification of bacteria and fungi, primarily due to the differences in their biological characteristics and genome structures. Bacterial identification predominantly employs the 16S rRNA gene sequencing technique. By sequencing and comparing the 16S rRNA gene found within bacterial cells, one can establish the phylogenetic position and taxonomic classification of bacteria. Fungi identification, on the other hand, primarily uses the ITS sequencing, motivated by the highly variable regions within the fungal genomes constituted by the ITS sequences.
Typically, bacteria possess three types of ribosomal RNA (rRNA): 5S rRNA (approximately 120bp), 16S rRNA (approximately 1540bp), and 23S rRNA (approximately 2900bp). Due to its short length and correspondingly limited genetic information, 5S rRNA gene sequences are ill-suited for the analysis and identification of bacterial species. Conversely, the 23S rRNA gene sequence is quite lengthy, and its high mutation rate does not favor the identification of bacteria with distant phylogenetic relations. The 16S rRNA, however, is ubiquitously found in prokaryotic cells, constituting over 80% of bacterial RNA due to its high abundance and copy number. This fact, coupled with its high functional homology and moderately sized genetic information, positions it as the benchmark for analysis of bacterial diversity. The 16S rRNA coding gene sequence includes ten conserved regions and nine highly variable regions. Among these, the V3-V4 region boasts excellent specificity and is well-represented in databases, rendering it the optimal choice for annotating analyses of bacterial diversity.
ITS reside between the fungal 18S, 5.8S, and 28S rRNA genes, with designated identifiers ITS 1 and ITS 2. In fungi, the 5.8S, 18S, and 28S rRNA genes display a high degree of preservation due to their essential role in protein synthesis, while the ITS regions, experiencing less evolutionary selection pressure, exhibit a higher tolerance for variation. As a result, ITS regions present considerable sequence polymorphism among fungi, especially within the eukaryotes. Interesting to note, the conserved nature of ITS sequences is manifested in a relative homogeneity intra-species while offering marked inter-species differentiation, enough to discern differences at the genus and even strain level. Bearing relatively short sequences, with lengths of around 350 bp and 400 bp for ITS 1 and ITS 2 respectively, ITS sequences are particularly amenable to analysis. This convenience, coupled with their polymorphic and differential properties, has led to the widespread use of ITS sequences in the phylogenetic analysis of various fungal taxa.
- Difference in sequencing applications
There are significant differences in the biological characteristics and lifestyles of bacteria and fungi, which directly influence their sequencing methodologies. Bacteria, typically unicellular microorganisms, possess a relatively smaller genome characterized by a high rate of replication and considerable adaptability. Thus, they are widely distributed in several environments, including soil, water bodies, the animal host, and more. Bacteria play diverse roles within ecosystems, comprising organic matter decomposition, nutrient production, symbiosis with plants and animals, among others. Consequently, the application of bacterial sequencing extends across various research fields such as environmental microbiomics, human microbiomics, and agricultural microbiology. In contrast, fungi are predominantly multicellular entities with relatively larger genomes and slower growth rates, and they demonstrate a relatively lower environmental adaptability. Fungi primarily survive by decomposing organic matter, playing the role of decomposers and biological degraders in nature. Thus, fungal sequencing finds more application in areas involving environmental microbiomics, disease diagnostics, and food safety, among other fields.
Bacteria sequencing applications:
1. Research on Microbial Diversity: This is typically accomplished through 16S rRNA gene sequencing of bacteria to amplify and sequence the bacterial 16S rRNA gene, subsequently rendering insight into the microbial community composition. Through this, we can understand the species composition, diversity indices, and community structure of microorganisms within a given sample. Such information contributes to a deepened comprehension of microbial diversity and functionality within varying environments, providing a scientific basis for ecosystem conservation and restoration.
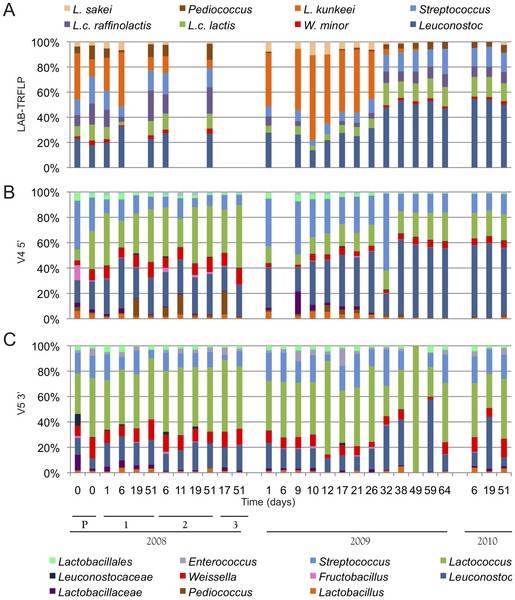 Figure 2. Sequencing to study bacterial diversity. (Bokulich et al., 2013)
Figure 2. Sequencing to study bacterial diversity. (Bokulich et al., 2013)
2. Pathogenic Bacterial Identification: Whole-genome sequencing and 16S rRNA gene sequencing are often used for pathogenic bacterial identification. Sequencing the genomic body of pathogens or certain specific genes enable precise identification of pathogenic microorganisms within samples. This can provide critical information for disease diagnosis and treatment by assisting in the development of targeted prevention and control measures, subsequently reducing the spread and outbreak of diseases and safeguarding public health.
3. Antimicrobial Resistance Analysis: Whole-genome sequencing or targeted gene sequencing is utilized for assessing bacterial resistance to antibiotics and other drugs. This process enables the detection of present antibiotic resistance genes and resistance mutations in the specimen, aiding in predicating the bacterial resilience level to different antibiotics. This approach facilitates the judicious use of antibiotics, minimizes bacterial drug resistance, and ensures the effectiveness of antibiotics.
4. Environmental Pollution Monitoring: Primarily, 16S rRNA gene sequencing is employed to analyze the microbiome structure within specimens, providing an assessment of environmental pollution levels. By detecting shifts in the composition of microbial communities within samples, we can evaluate environmental quality and the health status of ecosystems. This helps in promptly identifying and addressing environmental pollution issues, safeguarding ecological health and consequently, human wellbeing.
Fungi sequencing applications:
1. Diversity Research in Mycology: Primarily using Internal Transcribed Spacer (ITS) sequencing, information about the composition, abundance, and diversity of fungi in a sample can be obtained. This vital data aids in exploring the diversity and functions of fungi in different ecosystems, providing foundational information for research in fungal ecology.
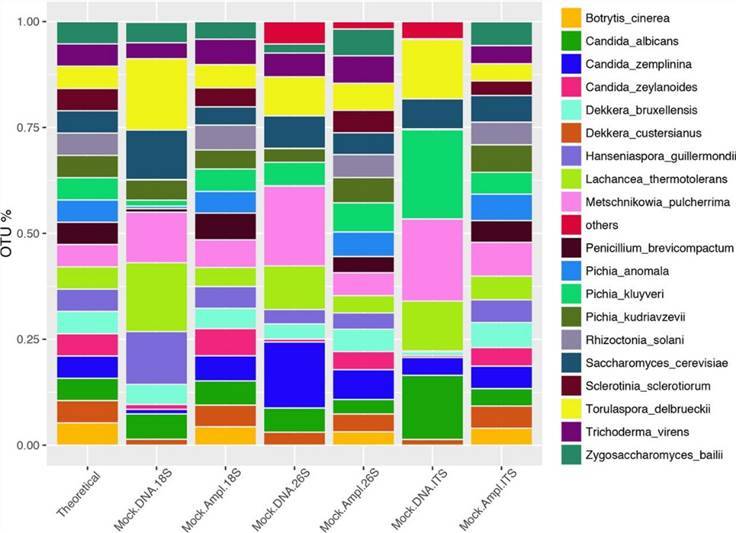 Figure 3. Bar graph of fungal community abundance. (De Filippis et al., 2017)
Figure 3. Bar graph of fungal community abundance. (De Filippis et al., 2017)
2. Identification of Fungal Pathogens: Using whole-genome sequencing and ITS sequencing, the genome and specific genes of pathogenic fungi can be analyzed. This allows for the rapid identification of pathogenic fungi in samples, determination of pathogenicity, and identification of transmission routes. This information is critical for precise diagnosis and disease prevention in the fields of medicine and crop protection, safeguarding human and crop health.
3. Biotechnological Applications: Incorporating whole-genome sequencing and transcriptomic sequencing to study fungal genomic structures and gene expression. This enables discovery and usage of fungal metabolic pathways and bioactive products, such as enzymes and antibiotics. This knowledge stimulates the development and utilization of fungal resources in the food and pharmaceutical industries, propelling industry innovation and economic advancement.
4. Biological Control and Bioremediation: Primarily using transcriptomic sequencing to study fungal metabolic pathways and gene regulation networks. This investigation uncovers key metabolic pathways and regulatory mechanisms of fungi in biological control and bioremediation. It offers innovative strategies and methods for environmental governance and ecological restoration, promoting the ecological recovery and health of soils, waters, and other environments.
CD Genomics is a genomics service company with a good reputation in providing reliable sequencing, genotyping, microarray, and bioinformatics services. Our fungal sequencing solutions vary from taxonomic profiling to discovering complex metabolic pathways to identifying metabolites that are beneficial to human health and more. Our platform provides data on the fungal genome sequence, helping to improve medical science, agriculture science, ecology, bioremediation, bioenergy, and biotech industries.
References
- Hibbett DS, Ohman A, Glotzer D, et al. Progress in molecular and morphological taxon discovery in Fungi and options for formal classification of environmental sequences. Fungal biology reviews. 2011, 25(1).
- Minerdi D, Bianciotto V, Bonfante P. Endosymbiotic bacteria in mycorrhizal fungi: from their morphology to genomic sequences. InDiversity and Integration in Mycorrhizas, 2002
- Li Q, Zhang B, He Z, et al. Distribution and diversity of bacteria and fungi colonization in stone monuments analyzed by high-throughput sequencing. PLoS One, 2016, 11(9): e0163287.
- Spini G, Spina F, Poli A, et al. Molecular and microbiological insights on the enrichment procedures for the isolation of petroleum degrading bacteria and fungi. Frontiers in Microbiology, 2018, 9: 2543.
- Alsohaili S A, Bani-Hasan B M. Morphological and molecular identification of fungi isolated from different environmental sources in the Northern Eastern desert of Jordan. Jordan Journal of Biological Sciences, 2018, 11(3).
- Bokulich NA, Joseph CML, Allen G, et al. Correction: Next-Generation Sequencing Reveals Significant Bacterial Diversity of Botrytized Wine. PLOS ONE, 2013, 8(10): 10.1371
- De Filippis F, Laiola M, Blaiotta G, et al. Different amplicon targets for sequencing-based studies of fungal diversity. Applied and Environmental Microbiology, 2017, 83(17): e00905-17.





 Figure 1. Aspergillums niger colony features on PDA (A1 Top, A2 Reverse) and conidia (A3). (Alsohaili et al., 2018)
Figure 1. Aspergillums niger colony features on PDA (A1 Top, A2 Reverse) and conidia (A3). (Alsohaili et al., 2018) Figure 2. Sequencing to study bacterial diversity. (Bokulich et al., 2013)
Figure 2. Sequencing to study bacterial diversity. (Bokulich et al., 2013) Figure 3. Bar graph of fungal community abundance. (De Filippis et al., 2017)
Figure 3. Bar graph of fungal community abundance. (De Filippis et al., 2017)