Introduction
Epigenetics, the discipline devoted to understanding heritable changes in gene expression independent of alterations to the DNA sequence, has emerged as a cornerstone in elucidating the intricate mechanisms dictating gene regulation. Among the diverse array of epigenetic modifications, methylation emerges as a central figure, exerting profound influence over gene expression profiles. This preamble serves to delve into the foundational tenets surrounding methylation, with particular emphasis on its critical role in epigenetic regulation. Moreover, we aim to delineate the essential disparities between RNA methylation and DNA methylation, emphasizing the imperative of distinguishing between these processes to achieve a comprehensive grasp of their individual contributions to gene regulation and cellular physiology. Through this discourse, our objective is to illuminate the pivotal significance of deciphering the complexities of methylation in shaping biological phenomena and delineating their ramifications in health and disease.
What is RNA methylation
RNA methylation entails the addition of methyl groups to particular nucleotides within RNA molecules, thereby exerting regulatory control over their functionality. Among the prevalent RNA methylation modifications, N6-methyladenosine (m6A) and 5-methylcytosine (5mC) stand out, with m6A being the most prevalent and thoroughly investigated. This modification predominantly occurs at adenosine residues, particularly within the consensus sequence RRACH (where R denotes purine, A signifies the m6A site, and H represents a non-guanine base).
Role of RNA methylation
RNA methylation constitutes a sophisticated, robust, and ever-adapting epigenetic alteration guiding gene expression, RNA stability, and the translation of proteins. It provides excellent specificity that enables fine-tuning of these fundamental biological processes. This modification imparts crucial regulatory control over integral aspects of RNA metabolism, hence influencing cellular phenotypic outcomes and functions. It also plays a significant role in safeguarding genomic integrity and directing the intricate mechanisms of broader biological systems.
Unraveling the intricate mechanisms of how RNA methylation modulates gene expression and protein translation could potentially reveal a wealth of untapped therapeutic targets. Such knowledge harbors immense prospective value in developing innovative strategic approaches for managing a multitude of human pathologies.
Regulating Gene Expression
RNA methylation dynamically regulates gene expression by modulating the abundance and activity of RNA transcripts. The addition of methyl groups to specific nucleotides within mRNA molecules exerts significant influence over various facets of mRNA metabolism, encompassing splicing, transport, and degradation. Notably, the m6A modification located within the 3' untranslated region (UTR) of mRNA serves to augment mRNA stability by shielding transcripts from degradation by ribonucleases. Furthermore, m6A modifications positioned within coding regions and 5' UTRs exert a nuanced control over translation efficiency, eliciting either enhancement or inhibition of protein synthesis contingent upon the contextual arrangement and distribution of methylation sites. Through meticulous modulation of mRNA transcript destiny, RNA methylation actively contributes to the precise orchestration of gene expression programs in response to developmental cues, environmental stimuli, and cellular signaling pathways.
Modulating RNA Stability
RNA methylation assumes a pivotal role in upholding the stability of RNA molecules, thus safeguarding the fidelity of genetic information transfer. The addition of methyl groups to RNA transcripts confers a shield against degradation by ribonucleases, thereby extending their half-life and bolstering their functional persistence within the cellular milieu. Furthermore, RNA methylation exerts precise control over the decay kinetics of select transcripts by modulating the recruitment of RNA-binding proteins and ribonucleoprotein complexes implicated in mRNA turnover pathways. Perturbations in the dynamic landscape of RNA methylation have been implicated in diverse pathological contexts, including cancer and neurodegenerative diseases, where dysregulated stability of RNA transcripts perpetuates disease progression and manifests phenotypic heterogeneity.
Orchestrating Protein Translation
RNA methylation intricately regulates protein translation by modulating both the efficiency and accuracy of ribosome-mediated synthesis of polypeptide chains. The presence of methylated nucleotides within mRNA transcripts influences the interplay between ribosomes and mRNA, thereby impacting translation initiation, elongation, and termination rates. Additionally, RNA methylation governs the accessibility of mRNA transcripts to ribosomal subunits and auxiliary translation factors, thereby modulating protein synthesis efficiency. Through precise adjustment of translational output for specific mRNA transcripts, RNA methylation dynamically contributes to the regulation of cellular proteome composition and function, critically influencing various physiological processes such as cell growth, differentiation, and homeostasis.
Mechanisms of RNA Methylation
As a molecular mechanism, RNA methylation embodies switch-like attributes that govern gene expression dynamics, via the orchestrated addition of methyl groups to specific nucleotides harbored within RNA molecules. This enzymatic interplay of events manipulates diverse RNA components - preferentially, the adenosine (A), cytosine (C), and occasionally including guanosine (G) and uridine (U) nucleotides.
Among these intricate modifications, m6A holds a notable position, being the most abundant and thoroughly researched facilitator of RNA methylation in eukaryotic cells. The adenosine residues' nitrogen-6 position undergoes methylation, driven by a complex synergized by pivotal enzymes such as methyltransferase-like 3 (METTL3) and methyltransferase-like 14 (METTL14).
These enzymes, intrinsic to the m6A machinery, serve as the arbiters of the RNA transcripts' fate by imprinting m6A marks, thereby directing the trajectory of multiple RNA metabolic operations and functionalities. Consequently, the systematic study of RNA methylation equips researchers with a comprehensive understanding of the modulation of cellular functionality, shaping potential therapeutic interventions for a plethora of human diseases.
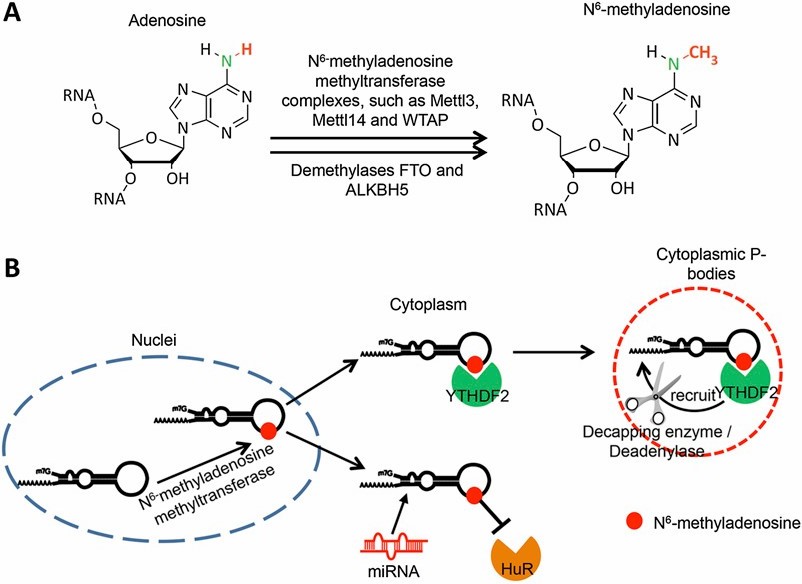 RNA methylation (m 6 A) and its potential biological functions.
RNA methylation (m 6 A) and its potential biological functions.
Enzymes Involved in RNA Methylation
RNA Methyltransferases: METTL3 and METTL14 stand as pivotal constituents within the m6A methyltransferase complex, pivotal in catalyzing the addition of methyl groups to adenosine residues nestled within RNA molecules. METTL3 assumes the role of the catalytic subunit, facilitating the transfer of the methyl group from S-adenosylmethionine (SAM) to the target adenosine. In parallel, METTL14 functions as an RNA-binding scaffold, amplifying substrate specificity and catalytic efficiency within the complex.
In concert with auxiliary proteins such as WTAP, VIRMA, and RBM15/15B, the METTL3-METTL14 duo orchestrates the site-specific deposition of m6A marks onto RNA transcripts, thus exercising precise control over their cellular fate and function.
Target Sites and Specificity in RNA Methylation
m6A Modification: The m6A modification predominantly manifests within the consensus motif DRACH (D = A/G/U, R = A/G, H = A/C/U), where the methylated adenosine residue (A) resides. Nonetheless, recent investigations have unveiled the occurrence of m6A deposition at non-canonical sites, thereby broadening the spectrum of potential target sites for RNA methylation. The spatial distribution of m6A marks across RNA transcripts exhibits dynamic variability contingent upon contextual factors, with notable enrichment observed in 3' untranslated regions (UTRs), proximal to stop codons, and within extensive internal exonic regions.
The precision of m6A deposition is governed by a multifaceted interplay involving sequence motifs, secondary RNA structures, and RNA-binding proteins that orchestrate the recruitment of the methyltransferase complex to specific loci within the transcriptome.
Other RNA Modifications:Beyond m6A, diverse forms of RNA methylation, including 5mC and N1-methyladenosine (m1A), undergo catalysis by discrete methyltransferases and display distinct preferences for target sites. For instance, 5mC modification predominantly localizes within CpG dinucleotides in DNA, whereas m1A modification exhibits enrichment within specific positions of tRNA molecules. The precise site specificity and functional ramifications of these RNA modifications remain under active investigation, with accumulating evidence underscoring their pivotal involvement in an array of cellular processes and disease etiology.
What is DNA methylation
DNA methylation, a covalent modification, entails the addition of a methyl group to the carbon-5 position of cytosine residues within DNA molecules. This biochemical process predominantly targets CpG dinucleotides, characterized by a cytosine followed by a guanine nucleotide. Through the methylation of CpG sites, DNA methylation emerges as a central regulator of gene expression dynamics, chromatin structure modulation, and the preservation of genomic integrity.
Role of DNA methylation
DNA methylation plays multifaceted roles in gene regulation, chromatin organization, genomic stability, developmental processes, and genomic imprinting. Its dynamic regulation and precise patterning are essential for normal cellular function and organismal development, with dysregulation of DNA methylation implicated in various human diseases.
Gene Regulation
DNA methylation constitutes a pivotal mechanism governing gene expression regulation. Methylation events targeting CpG dinucleotides within gene promoter regions frequently instigate transcriptional repression by impeding the binding of transcription factors and facilitating the recruitment of methyl-binding proteins, thereby instigating chromatin compaction and impeding access of the transcriptional machinery to the gene promoter. Conversely, hypomethylation within promoter regions correlates with heightened gene transcription activity, facilitating the recruitment of transcription factors and RNA polymerase for robust gene expression.
Chromatin Structure
Complementarily, the methylation of DNA influences the structure and composition of chromatin, thus playing a defining role in gene expression. CpG sites that have been methylated are correlated with a compacted chromatin structure, designated as heterochromatin, which accordingly, results in the transcriptional muting of the related genes. The resultant heterochromatin state obstructs the binding of transcriptional activators and promotes the engagement of histone modifiers such as histone deacetylases and methyltransferases, fortifying the repressive chromatin habitat.
Contrarily, regions experiencing hypomethylation are related to an open chromatin conformation, thereby facilitating active gene expression. These integral biochemical modifications underscore the power of methylation and its significance in gene regulation.
Genomic Stability
DNA methylation serves as an indispensable determinant in upholding genomic stability through the regulation of repetitive DNA constituents such as transposable elements and retroviruses. Enforced through methylation, the immobility of such repetitive elements effectively curbs their erratic reactivation, constituting an essential surveillance mechanism for genomic integrity. Conversely, the relinquishment of DNA methylation within repetitive sequences may precipitate genomic instability, chromosomal rearrangements, and the reactivation of retrotransposons. This cascade of events potentially underpins the etiology of a multitude of diseases, notably encompassing malignancies and developmental aberrations.
Developmental Processes
DNA methylation constitutes a fundamental aspect of normal development and cellular differentiation. Throughout embryonic development, dynamic alterations in DNA methylation patterns undergo stringent regulation to coordinate lineage-specific gene expression programs. These patterns are established and upheld by DNA methyltransferases during the developmental trajectory, thereby ensuring precise gene regulation and delineation of cell fate. Deviations from the normative DNA methylation landscape during development can precipitate developmental anomalies and disease states.
Genomic Imprinting
DNA methylation intricately participates in genomic imprinting, an epigenetic phenomenon whereby specific genes exhibit parent-of-origin-specific expression patterns. Imprinted genes frequently harbor differentially methylated regions (DMRs) that undergo parent-specific DNA methylation events during gametogenesis. The establishment of DNA methylation at these DMRs governs allele-specific gene expression, thereby engendering parentally biased expression profiles pivotal for normal developmental processes and growth.
Mechanisms of DNA methylation
DNA methylation, an integral locus of epigenetic control, has crucial functions involving two primary mechanisms: maintenance and de novo methylation. Maintenance methylation, efficiently catalysed by DNA methyltransferase 1 (DNMT1), perpetuates the segregation of established DNA methylation patterns during cell replication. It specifically targets hemimethylated DNA strands that result from DNA replication, thereby ensuring the meticulous conservation of epigenetic signatures on nascent DNA strands.
In contrast, de novo methylation, an intricate process conducted primarily by DNMT3A and DNMT3B, instigates methylation patterns onto CpG sites previously unmethylated. This specific phenomenon underpins pivotal biological processes such as embryogenesis, cellular differentiation, and adaptive responses to environmental stimuli.
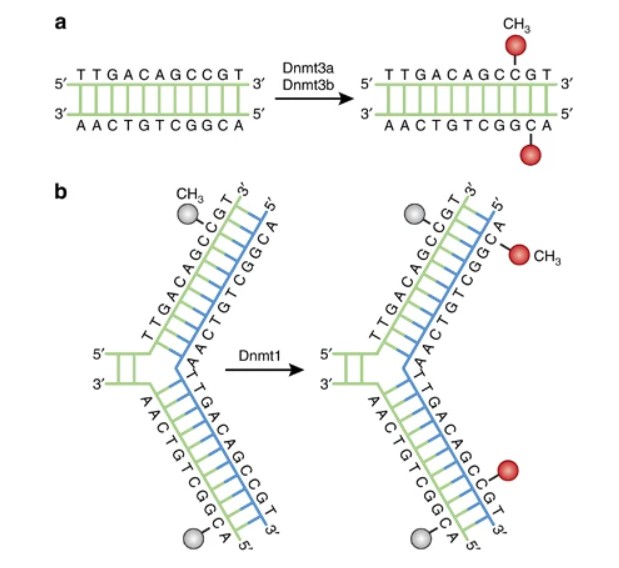 DNA methylation pathways
DNA methylation pathways
Enzymes of DNA Methylation
Enzymes of DNA methylation play a pivotal role in catalyzing the addition of methyl groups to cytosine residues in DNA molecules. These enzymes are primarily responsible for establishing and maintaining DNA methylation patterns, thereby regulating gene expression and chromatin structure.
DNA Methyltransferases (DNMTs):
DNMT1, the predominant DNA methyltransferase in mammalian cells, assumes a pivotal role in the maintenance of DNA methylation patterns throughout DNA replication. It discerns hemi-methylated CpG sites, where one DNA strand is methylated, and proceeds to methylate the nascently synthesized strand, thereby preserving methylation patterns across successive cell divisions.
In contrast, DNMT3A and DNMT3B function as de novo DNA methyltransferases, orchestrating the establishment of fresh DNA methylation patterns during early developmental stages and cellular differentiation processes. Integral to the creation of tissue-specific methylation profiles, DNMT3A and DNMT3B serve critical roles in embryonic development, genomic imprinting, and the process of X-chromosome inactivation.
Ten-Eleven Translocation (TET) Proteins:
TET proteins, encompassing TET1, TET2, and TET3, function as dioxygenases, catalyzing the conversion of 5mC to 5hmC, thereby initiating the process of active DNA demethylation. Via iterative oxidation reactions, TET proteins further catalyze the transformation of 5hmC into 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC), culminating in DNA demethylation mediated by base excision repair mechanisms.
The orchestrated action of TET-mediated DNA demethylation is indispensable for the dynamic regulation of DNA methylation patterns throughout processes such as development, cellular differentiation, and the response to environmental cues. Perturbations in TET protein function have been implicated in a spectrum of diseases, including cancer and neurological disorders.
UHRF1 (Ubiquitin-like with PHD and RING Finger domains 1):
UHRF1 emerges as a pivotal orchestrator in the meticulous preservation of DNA methylation, exerting indispensable control over the recruitment of DNMT1 to hemi-methylated CpG loci during the process of DNA replication. This regulatory prowess hinges on its adept recognition of hemi-methylated CpG sites, a faculty executed through its SRA (SET and RING associated) domain. By virtue of this recognition mechanism, UHRF1 ensures the precise allocation of DNMT1, thereby safeguarding the fidelity of DNA methylation patterns.
Beyond its role in DNA methylation maintenance, UHRF1 manifests an additional facet of functionality, wielding E3 ubiquitin ligase activity. This catalytic capability facilitates the ubiquitination and subsequent degradation of histone H3 lysine 9 (H3K9) methyltransferases. Through this enzymatic cascade, UHRF1 fosters the induction of heterochromatin formation and engenders transcriptional repression at methylated CpG sites. Such multifaceted actions underscore the nuanced regulatory landscape governed by UHRF1, wherein it orchestrates intricate epigenetic processes critical for cellular homeostasis.
Other Regulatory Proteins:
Supplementary proteins, exemplified by MeCP2 (Methyl-CpG-binding protein 2) and MBD proteins (Methyl-CpG-binding domain proteins), emerge as pivotal participants in deciphering and disseminating DNA methylation cues. MeCP2 exhibits a targeted affinity for methylated CpG dinucleotides, thereby facilitating the recruitment of histone deacetylases and chromatin remodeling complexes. This concerted action precipitates transcriptional repression, thus modulating gene expression dynamics.
Similarly, the cadre of MBD proteins, encompassing MBD1, MBD2, MBD3, and MBD4, assumes roles of significance in the epigenetic landscape. These proteins exhibit a capacity to discern methylated CpG sites, thereby serving as conduits for orchestrating transcriptional repression or engaging in DNA repair processes. Through these molecular engagements, MeCP2 and MBD proteins intricately weave into the regulatory tapestry governing gene expression and genomic integrity, underscoring their indispensability in cellular physiology.
Interplay Between RNA and DNA Methylation
The dynamic regulatory interrelation between DNA and RNA methylation orchestrates transcriptional processes and cellular functionality. Distinctly pivotal roles are embodied by the reciprocal and interconnected nature of these two epigenetic modifications in determining the course of cellular events and in safeguarding genomic stability.
Scrutinizing the intricate dialogues between RNA and DNA methylation uncovers complex regulatory systems governing gene expression and chromatin structuring. RNA methylation, with significant emphasis on m6A modification, is now understood to be a crucial contributor in the post-transcriptional gene regulatory environment. Evidence clearly demonstrates the potential of RNA methyltransferases, including but not limited to METTL3 and METTL14, to dynamically alter mRNA transcripts, consequently affecting mRNA stability, splicing processes, and translation efficiency. Of notable import is the ability of these RNA methylation occurrences to shape DNA methylation landscapes by influencing the expression of DNA methyltransferases and chromatin remodelers.
In parallel, DNA methylation can exercise regulatory force over RNA methylation dynamics. Methylation of DNA regions is linked with changes in chromatin conformation and transcriptional silencing, which subsequently result in the attraction of chromatin-modifying enzymes capable of influencing RNA methylation mechanisms. For instance, DNA methylation situated at gene promoters can obstruct connectivity between transcription factors and RNA polymerase, thereby influencing both the elongation and processing of emergent RNA transcripts. Additionally, DNA methylation-led silencing of non-coding RNAs, inclusive of microRNAs and long non-coding RNAs, can indirectly modulate RNA methylation landscapes through alterations in the availability of RNA methylation regulators.
Multiple regulatory pathways illuminate the complex interconnections between DNA and RNA methylation processes. For instance, epigenetic modifications of enhancer locales, encompassing both DNA methylation and histone modifications, can influence the transcription of enhancer RNAs (eRNAs), which, in turn, affect chromatin accessibility and gene expression values. Correspondingly, the governance exerted by RNA methylation on RNA processing and stability can impact the expression and activity of DNA methylation regulators, thereby fostering feedback cycles critical to sustaining epigenetic states.
The complex synergy between RNA and DNA methylation bears significant ramifications on the modulation of gene expression and cellular activity. Unbalances in this dynamic interaction have been associated with multiple pathological conditions, including but not restricted to cancer, neurological syndromes, and developmental anomalies. Decoding the intertwined mechanisms of RNA and DNA methylation is critical in understanding the molecular foundations of disease pathogenesis and in the identification of potential therapeutic targets. Moreover, elucidating the functional outcomes arising from RNA-DNA methylation discourse may lead to novel diagnostic biomarkers and prognostic indices pertaining to disease progression and treatment responses.
Overview of Cutting-Edge Techniques for Studying RNA methylation
In the dynamic realm of RNA methylation research, a plethora of cutting-edge technologies have emerged to dissect the intricate landscape of epitranscriptomics. From high-throughput sequencing methods to innovative biochemical assays, these techniques offer unprecedented insights into the mechanisms and functional implications of RNA methylation. Below, we delve into the specifics of each method and present them in a concise tabular format for easy reference.
| Technology |
Description |
Application |
| Methylated RNA Immunoprecipitation Sequencing (MeRIP Sequencing) |
Antibody-based immunoprecipitation enriches methylated RNA fragments, followed by high-throughput sequencing for profiling RNA methylation patterns. |
Comprehensive mapping of methylated RNA sites across the transcriptome. |
| Bisulfite Sequencing |
Chemical treatment converts unmethylated cytosines to uracil while leaving methylated cytosines intact, enabling single-base resolution mapping of RNA methylation. |
Identification of methylated cytosines within RNA transcripts. |
| RNA Methylation-Sensitive Enzymes and Probes |
Enzymatic and chemical approaches selectively target methylated RNA residues, enabling site-specific mapping and quantification of RNA methylation. |
Probing RNA methylation dynamics and interactions within RNA-protein networks. |
| High-Throughput Screening Assays |
Large-scale screening assays facilitate the discovery of novel RNA methyltransferases, demethylases, and reader proteins involved in RNA methylation dynamics. |
Systematic evaluation of genetic and chemical perturbations on RNA methylation pathways. |
| NanoPore Sequencing with Direct RNA Sequencing |
Direct detection of RNA/DNA modifications, including RNA methylation, using nanopore sequencing technology. |
Real-time detection of RNA modifications without the need for prior bisulfite treatment or reverse transcription. |
Advancements in DNA Methylation Study Technologies
DNA methylation study technologies have undergone remarkable advancements, revolutionizing our ability to interrogate epigenetic landscapes with precision and throughput. By unraveling the complexities of DNA methylation mechanisms and leveraging innovative methodologies, researchers at Creative Proteomics and beyond are poised to unlock new insights into the role of DNA methylation in health, disease, and beyond.
| Technology |
Description |
Application |
| Whole Genome Bisulfite Sequencing (WGBS) |
Bisulfite treatment converts unmethylated cytosines to uracil, allowing differential methylation analysis at single-base resolution. |
Comprehensive mapping of DNA methylation patterns across the entire genome. |
| Reduced Representation Bisulfite Sequencing (RRBS) |
Selective digestion with methylation-insensitive restriction enzymes followed by bisulfite treatment enables methylation profiling of CpG-rich regions. |
Cost-effective approach for analyzing DNA methylation patterns in CpG islands and gene regulatory regions. |
| Methylated DNA Immunoprecipitation Sequencing (MeDIP-Seq) |
Antibody-based enrichment of methylated DNA fragments followed by high-throughput sequencing for profiling DNA methylation patterns. |
Identification of methylated regions in the genome with relatively low sequencing depth. |
| Methyl-CpG Binding Domain Sequencing (MBD-Seq) |
Methyl-CpG binding domain proteins are used to capture methylated DNA fragments, followed by sequencing for DNA methylation analysis. |
Enrichment of methylated DNA regions for genome-wide DNA methylation profiling. |
| Methylation-Sensitive Restriction Enzyme Sequencing (MRE-Seq) |
Digestion of genomic DNA with methylation-sensitive restriction enzymes followed by sequencing to identify methylated CpG sites. |
Detection of DNA methylation patterns at specific recognition sites. |
| Targeted Bisulfite Sequencing |
Customized primer design targets specific genomic regions for bisulfite treatment and sequencing, providing focused analysis of DNA methylation. |
Profiling methylation status in regions of interest with high coverage and resolution. |
| |
|
|
| Methylation-Specific PCR (MSP) |
PCR amplification using primers specific to methylated or unmethylated DNA, followed by gel electrophoresis for methylation status determination. |
Rapid and cost-effective detection of DNA methylation at specific CpG sites. |
| Pyrosequencing |
Quantitative analysis of DNA methylation levels at single-base resolution using sequencing-by-synthesis technology. |
Accurate measurement of methylation levels in CpG dinucleotides, suitable for small-scale DNA methylation studies. |
| Methylation Array |
Microarray-based assay for genome-wide detection of DNA methylation patterns, utilizing probes specific to methylated CpG sites. |
High-throughput screening of DNA methylation status across thousands of CpG sites simultaneously. |
| CRISPR-based Epigenome Editing |
CRISPR-Cas9 technology engineered to modulate DNA methylation patterns by targeting specific genomic loci. |
Functional validation of DNA methylation-mediated gene regulation and epigenetic modifications. |
| Single-cell DNA Methylation Sequencing |
Single-cell resolution sequencing techniques enabling the analysis of DNA methylation heterogeneity within individual cells. |
Investigation of cell-to-cell variability in DNA methylation patterns and its role in cellular function and differentiation. |
Summary of RNA methylation vs DNA methylation
| Aspect |
RNA Methylation |
DNA Methylation |
Shared Aspects |
| Definition |
Covalent modifications of RNA molecules, including m6A, m5C, and Ψ. |
Covalent addition of a methyl group to cytosine residues in DNA molecules, primarily at CpG dinucleotides. |
Covalent modifications of nucleic acids. |
| Target Residues |
Primarily targets adenine (m6A), cytosine (m5C), and uridine (Ψ) residues in RNA. |
Targets cytosine residues in DNA, predominantly at CpG dinucleotides. |
Methylation occurs at specific nucleotide residues. |
| Enzymes Involved |
RNA methyltransferases, demethylases, and reader proteins. |
DNA methyltransferases (DNMTs) - DNMT1, DNMT3A, DNMT3B; Ten-eleven translocation (TET) enzymes. |
Involvement of methyltransferases and demethylases. |
| Role in Gene Expression |
Regulates mRNA stability, splicing, translation, and RNA metabolism. |
Regulates transcriptional activity, chromatin compaction, and gene silencing. |
Influence on gene expression and cellular processes. |
| Regulatory Mechanisms |
Influences post-transcriptional gene regulation. |
Governs transcriptional repression and chromatin structure. |
Regulation of gene expression through epigenetic modifications. |
| Biological Functions |
Essential for cell differentiation, development, and response to environmental cues. |
Maintains genomic stability, cellular identity, and cellular homeostasis. |
Critical roles in cellular processes and organismal development. |
| Implications in Disease |
Dysregulation linked to cancer, neurodegenerative diseases, and metabolic disorders. |
Associated with cancer, developmental disorders, and autoimmune diseases. |
Implicated in various diseases and pathological conditions. |
| Interplay |
Crosstalk with DNA methylation and other epigenetic modifications. |
Interacts with histone modifications and chromatin remodeling complexes. |
Interconnected regulatory networks in epigenetics. |
Conclusion
To conclude, a comparative examination of RNA and DNA methylation elicits their distinct characteristics and communal regulatory responsibilities in cellular operations. DNA methylation predominantly focuses on the covalent alteration of cytosine deposits at CpG dinucleotides, whereas RNA methylation involves a plethora of modifications, inclusive of m6A, m5C, and Ψ motifs. Irrespective of their structural divergence, both RNA and DNA methylation constitute integral roles in dictating genetic expression blueprints, chromatin conformation, and cellular operation.
Specifically, RNA methylation, with particular focus on m6A modification, has emerged as a principal factor in post-transcriptional regulation, impacting mRNA constancy, splicing, and translation efficacy. In contrast, DNA methylation chiefly directs transcriptional suppression and chromatin condensation, contributing to the genesis and perpetuation of cellular individuality and genomic robustness. Both epigenetic alterations intersect at diverse regulatory junctions, architecting elaborate networks that fine-tune genetic expression and cellular reactions to ecological stimuli.
Cognizance of the divergences and commonalities between RNA methylation and DNA methylation is integral for propelling biomedical investigation and for therapeutic innovation. Abnormalities in such epigenetic changes, featuring anomalous RNA and DNA methylation profiles, have been associated with a variety of human maladies including cancer, neurodegenerative conditions, and metabolic disorders. Therefore, demystifying the molecular machinations that underlie RNA-DNA methylation intercommunication offers potential for the identification of novel diagnostic indicators and therapeutic objectives for precision medicine approaches.
In summary, the exploration of RNA and DNA methylation constitutes a swiftly growing discipline with immense promise for unraveling the convolutions of genetic regulation and cellular physiology. Through enlightening the interactions between these epigenetic modifications, investigators can deepen their understanding of disease genesis and devise innovative diagnostics, prognostic strategies, and therapeutics. Ultimately, the encompassing comprehension of RNA-DNA methylation dynamics holds potential to catalyze transformative progression in the realms of biomedical research and personalized medicine.
References:
-
Moore, L., Le, T. & Fan, G. DNA Methylation and Its Basic Function. Neuropsychopharmacol 38, 23–38 (2013).
- Kurdyukov S, Bullock M. DNA Methylation Analysis: Choosing the Right Method. Biology (Basel). 2016
- Jin B, Li Y, Robertson KD. DNA methylation: superior or subordinate in the epigenetic hierarchy? Genes Cancer. 2011
- Dor Y, Cedar H. Principles of DNA methylation and their implications for biology and medicine. Lancet. 2018
- Boulias, K., Greer, E.L. Biological roles of adenine methylation in RNA. Nat Rev Genet 24, 143–160 (2023).
- Yao, B., & Jin, P. (2014). Unlocking epigenetic codes in neurogenesis. Genes & Development, 28, 1253 - 1271.


 Sample Submission Guidelines
Sample Submission Guidelines
 RNA methylation (m 6 A) and its potential biological functions.
RNA methylation (m 6 A) and its potential biological functions. DNA methylation pathways
DNA methylation pathways