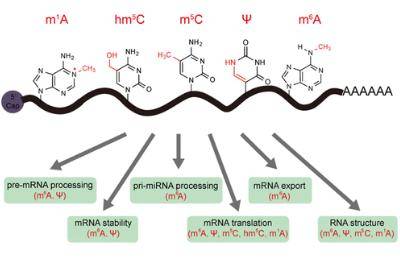
What Is RNA Modifications
RNA modifications play a pivotal role in the epigenetic regulation of post-transcriptional gene expression. While extensive research has been conducted on m6A RNA modifications, various other types of RNA modifications also participate in the regulation of post-transcriptional gene expression. These include m1A, m5C, m7G, 2'-O-methylation, and ac4C acetylation. Notably, ongoing studies in these areas continue to yield high-impact publications.
CD Genomics stands as a leader in RNA modification sequencing, offering a comprehensive suite of RNA modification sequencing services tailored for mRNA and various non-coding RNAs. Our services cover a spectrum of RNA modifications, including m6A, m1A, m5C, m7G, ac4C, 2'-O-methylation, and pseudouridine (Ψ), contributing to advancing the understanding of the intricate landscape of RNA modifications.
 (Jinghui Song et al,. ACS Chem. Biol. 2017)
(Jinghui Song et al,. ACS Chem. Biol. 2017)
RNA Modification Detction Methods
(1) m6A RNA Methylation Sequencing: Sensitive Techniques Simultaneously Detecting m6A Modifications in mRNA, lncRNA, and Circular RNA
Quantitative and Trace-Level m6A RNA Methylation Sequencing (MeRIP/m6A-seq)
m6A methylation is a widespread RNA modification in eukaryotic organisms, including mRNA and even viral RNA. Although m6A modification was reported as early as the 1970s, comprehensive knowledge about its overall distribution in RNA and its impact on gene expression regulation has been limited. The discovery of the first bona fide RNA demethylase, FTO, in 2011 renewed interest in the methylation of mRNA/lncRNA.
MeRIP, employing m6A-specific antibodies for enrichment followed by sequencing, is a method designed to investigate adenosine methylation modifications in RNA. EasyGene has independently developed a trace-level RNA methylation detection technology, enabling the reduction of sample input to 10-20 μg, with a minimum requirement of only 5 μg of total RNA. This advancement facilitates precise analysis of m6A modifications across various RNA species, including mRNA, lncRNA, and circular RNA.
(2) m5C RNA Methylation Sequencing: Bisulfite Treatment for Single-Base Precision Detection of m5C Modification Levels
m5C RNA Methylation (RNA-BS-seq, m5C)
Among the numerous RNA modifications, m5C is one extensively studied. When present in tRNA, m5C can regulate translation; on rRNA, it plays a role in quality control during ribosome biogenesis, and when found on mRNA, it influences mRNA structure, stability, and translation processes.
RNA-BS-seq stands out as a powerful technique for detecting m5C. This method involves treating RNA with bisulfite, converting unmodified cytosines (C) to uracils (U), while m5C-modified cytosines remain unaltered. Through subsequent PCR, uracils are converted to thymines (T), allowing the discrimination between m5C and C. Coupled with high-throughput sequencing, RNA-BS-seq enables the comprehensive detection of m5C modifications across the entire transcriptome.
(3) RIP-seq: Unraveling RNA-Protein Interactions through Immunoprecipitation-Based Sequencing
RNA Immunoprecipitation sequencing (RIP-seq) stands as a robust methodology employed to delve into the intricate landscape of RNA-protein interactions, offering insights into the dynamic associations between RNA molecules and their interacting proteins. This technique hinges on the precise capture of RNA-protein complexes through immunoprecipitation, facilitated by antibodies directed against the protein of interest.
Wielding considerable significance in the exploration of post-transcriptional gene regulation, RIP-seq serves as a pivotal tool for identifying RNA targets bound by specific proteins. Post immunoprecipitation, the associated RNA is meticulously isolated and subjected to high-throughput sequencing, furnishing a comprehensive profile of RNA molecules actively participating in protein interactions. This approach plays a pivotal role in advancing our comprehension of fundamental cellular processes, encompassing RNA processing, localization, and stability.
(4) Nanopore RNA Methylation Sequencing Service: Revealing the Fine Details of RNA Modification Patterns with Single-Nucleotide Precision
Our Nanopore RNA Methylation Sequencing Service represents an advanced solution tailored to unveil the nuanced landscape of RNA modifications with unparalleled precision. Harnessing the capabilities of nanopore sequencing technology, this service delivers single-nucleotide resolution, facilitating the identification and characterization of diverse RNA modifications, including m6A, m1A, m5C, m7G, ac4C, and 2'-O-methylation.
Distinguished by its capacity to capture subtle nuances in RNA modifications across various RNA species, encompassing mRNA and non-coding RNA, this service utilizes nanopore technology for real-time, long-read sequencing. This approach ensures the detection of modifications with heightened accuracy and sensitivity. Emphasizing comprehensive and high-throughput analysis, our service empowers researchers to delve into the dynamic realm of RNA modifications, unraveling their functional implications with profound insights.
(5) 2'-O-Methylated-seq: Profiling 2'-O-Methylation Patterns in RNA Molecules
Our 2'-O-Methylated-seq service is dedicated to elucidating the intricate patterns of 2'-O-methylation in RNA molecules. This modification, occurring at the 2'-hydroxyl group of ribose, plays a pivotal role in RNA stability and function. Leveraging state-of-the-art sequencing technologies, this service delivers high-resolution data on the distribution of 2'-O-methylation across diverse RNA species, encompassing mRNA, lncRNA, pri-miRNA, tRNA, and rRNA.
Prioritizing both accuracy and efficiency, 2'-O-Methylated-seq provides valuable insights into the specific locations and abundance of 2'-O-methylation sites within RNA molecules. The service excels in handling a variety of sample types, ensuring comprehensive coverage and robust detection of 2'-O-methylation events. Researchers can rely on this service to explore the nuanced realm of 2'-O-methylation, shedding light on its functional relevance and potential implications in biological processes.
acRIP-seq: High-Throughput Profiling of Acetylated RNA Regions
Acetylated RNA Immunoprecipitation Sequencing (acRIP-seq) represents a pioneering approach to delineate RNA acetylation patterns across the transcriptome. This technique harnesses antibodies that specifically bind to acetylated nucleotide bases, facilitating the immunoprecipitation-mediated enrichment of acetylated RNA fragments. Subsequent high-throughput sequencing permits a comprehensive analysis of the distribution and frequency of acetylated sites in various RNA species. Acetylation, notably N4-acetylcytidine (ac4C), has been identified as a crucial modification influencing protein translation, RNA stability, and alternative splicing. Recent investigations have revealed that ac4C, previously thought to be predominantly associated with tRNA and 18S rRNA, is abundantly present in other RNA types. These findings underscore the significant role of ac4C in the regulation of gene expression and highlight its potential as a pivotal element in the field of epitranscriptomics.
PA-Ψ-seq: Unraveling Pseudouridine Modifications with Single-Nucleotide Precision
Photo-crosslinking-assisted Ψ sequencing (PA-Ψ-seq) is a sophisticated methodology designed to achieve high-resolution mapping of pseudouridine (Ψ) modifications at the single-nucleotide level. This technique employs photo-crosslinking to stabilize Ψ-modified RNA, followed by antibody-mediated enrichment and high-throughput sequencing. PA-Ψ-seq excels in generating comprehensive pseudouridine modification maps across the entire transcriptome, providing valuable insights into the role of Ψ modifications in various biological processes, including gene regulation and disease. While the functional significance of Ψ in rRNA and tRNA is well established, its role in mRNA has remained elusive due to technical challenges in detection. PA-Ψ-seq addresses these limitations, offering a robust tool for elucidating the functional implications of Ψ modifications in mRNA and examining their potential alterations in pathological conditions, such as cancer.
Here are some publications that have been successfully published using our services or other related services:
Restriction endonuclease cleavage of phage DNA enables resuscitation from Cas13-induced bacterial dormancy
Journal: Nature microbiology
Year: 2023
IL-4 drives exhaustion of CD8+ CART cells
Journal: Nature Communications
Year: 2024
High-Fat Diets Fed during Pregnancy Cause Changes to Pancreatic Tissue DNA Methylation and Protein Expression in the Offspring: A Multi-Omics Approach
Journal: International Journal of Molecular Sciences
Year: 2024
KMT2A associates with PHF5A-PHF14-HMG20A-RAI1 subcomplex in pancreatic cancer stem cells and epigenetically regulates their characteristics
Journal: Nature communications
Year: 2023
Cancer-associated DNA hypermethylation of Polycomb targets requires DNMT3A dual recognition of histone H2AK119 ubiquitination and the nucleosome acidic patch
Journal: Science Advances
Year: 2024
Genomic imprinting-like monoallelic paternal expression determines sex of channel catfish
Journal: Science Advances
Year: 2022
See more articles published by our clients.


 Sample Submission Guidelines
Sample Submission Guidelines
