Single-cell T cell receptor sequencing (scTCR Seq) represents a pioneering advancement in the study of immune responses. This technology enables the analysis of TCRs at the single-cell level, providing a detailed view of T cell diversity and specificity.. Such detailed insights are critical for advancing research in oncology, autoimmune disorders, and the development of personalized immunotherapeutics. This article delves into the importance, applications, and benefits of scTCR Seq, elucidating its transformative impact on the field of immunology.
What is scTCR Seq?
scTCR Seq is a sophisticated methodological advancement that enables researchers to scrutinize the T cell receptor (TCR) repertoire with unparalleled single-cell resolution. This approach effectively captures the intricate diversity and specificity of T cell responses across a range of biological contexts, including malignancies, infectious agents, and autoimmune disorders. Through the isolation and sequencing of individual T cells, scTCR Seq facilitates a profound comprehension of immune system operations at the cellular level.
Why is it Important?
Conventional TCR sequencing methodologies generally evaluate TCRs derived from bulk T cell populations, yielding a generalized overview of receptor diversity. In contrast, scTCR Seq provides precise and comprehensive data regarding the receptor profiles of individual T cells, encompassing both TCR alpha and beta chains. This level of detail affords significant insights into T cell clonality and functional responses, which are vital for the progression of personalized medical approaches and the refinement of cancer immunotherapies.
For further insights into TCR sequencing and its contributions to immunological research, interested readers are encouraged to explore our dedicated page on TCR Sequencing Services.
Advantages of scTCR Seq Over Traditional TCR Sequencing
Paired TCR Chain Sequencing
scTCR-seq sequencing offers precise identification of T-cell clonotypes, enabling researchers to accurately determine the α and β chain pairings within individual T cells. This ability is crucial for understanding T cell specificity and function. Stubbington et al. exemplified the efficacy of paired TCR sequencing in pinpointing antigen-specific T cells within the context of a Salmonella infection study.
Integration with Transcriptomics
The integration of scTCR-seq with transcriptomic profiling facilitates the linkage of TCR sequences to gene expression patterns, yielding valuable insights into T cell functionality. This correlation provides a holistic perspective on T cell biology. Zhang et al. successfully employed this methodology to delineate distinct transcriptional signatures related to varied TCR clonotypes in tumor-infiltrating lymphocytes.
High-Throughput Capability
The high-throughput nature of modern scTCR-seq platforms allows for the concurrent analysis of thousands of cells within a single experimental setting. This capability is pivotal for comprehensive profiling of T cell populations. Zheng et al.'s work, utilizing a droplet-based approach, highlighted the capacity to process over 5,000 single T cells in a solitary experiment.
High Resolution
ScTCR-seq delivers exceptional resolution in the investigation of T-cell heterogeneity, providing insights into the diversity and clonal expansion of T cells at the single-cell level. This high-resolution analysis was showcased in a study by Guo et al., which unveiled the clonal architecture of tumor-infiltrating T cells in colorectal cancer.
Clonal Analysis
The precise pairing of TCR α and β chains enabled by scTCR-seq is integral to understanding immune responses. Wu et al. harnessed this capability to examine 47 tumor biopsies from patients with non-small cell lung cancer, uncovering critical insights into T cell dynamics during immunotherapy.
Functionality Linkage
This technique enables the association of T cell functions—such as cytotoxicity and cytokine production—with specific TCRs. In melanoma patients undergoing checkpoint blockade therapy, Yost et al. utilized scTCR-seq to trace T cell clones, thereby illustrating the relationship between clonal expansion and functional phenotypes.
Enhanced Immunotherapy Design
ScTCR-seq offers pivotal insights critical for designing targeted immunotherapies. Penter et al.'s application of scTCR-seq to analyze CAR-T cell products revealed vital information regarding their clonal composition and potential efficacy, providing guidance for the development of more effective cellular therapies. This cutting-edge tool is revolutionizing research across domains including immunotherapy, vaccine development, and transplant medicine. Notably, Yost et al.'s use of scTCR-seq in monitoring the dynamics of individual T cell clones during checkpoint blockade therapy in cancer immunotherapy has elucidated mechanisms underpinning treatment responses and resistance.
With these groundbreaking capabilities, scTCR-seq is paving the way for new discoveries in immunological research and therapy development.
How scTCR Seq Works
The scTCR sequencing process, while intricate, offers profound insights into immune responses by elucidating the detailed profiles of T cell receptors. Below is a systematic overview of the key steps involved:
Cell Isolation
The initiation of scTCR sequencing involves the isolation of individual T cells, a critical step achieved through advanced techniques such as microfluidics or Fluorescence-Activated Cell Sorting (FACS). This step ensures the compartmentalized processing of each T cell, maintaining the integrity and specificity of subsequent analyses.
TCR Amplification
Subsequent to isolation, TCR genes, encompassing both alpha and beta chains, undergo amplification via Polymerase Chain Reaction (PCR). This amplification is crucial for generating a sufficient quantity of genetic material conducive to sequencing, facilitating the detailed study of TCRs.
Sequencing
The amplified TCR sequences are subjected to Next-Generation Sequencing (NGS) technologies. Through this step, researchers can accurately identify and characterize the distinct TCRs present within individual T cells, enabling a comprehensive understanding of T cell receptor diversity and specificity.
Data Analysis
In the final stage, sophisticated bioinformatics tools are employed to process and analyze the vast corpus of sequencing data generated. This computational analysis aids in the pairing of TCR alpha and beta chains and the identification of T cell clones, providing invaluable insights into the immune repertoire.
The scTCR sequencing workflow is highly detailed and necessitates the use of state-of-the-art technologies.
For an in-depth examination of TCR profiling strategies, refer to our article on TCR Detection Methods. This exploration highlights the versatility and precision of scTCR sequencing in advancing our understanding of immune responses.
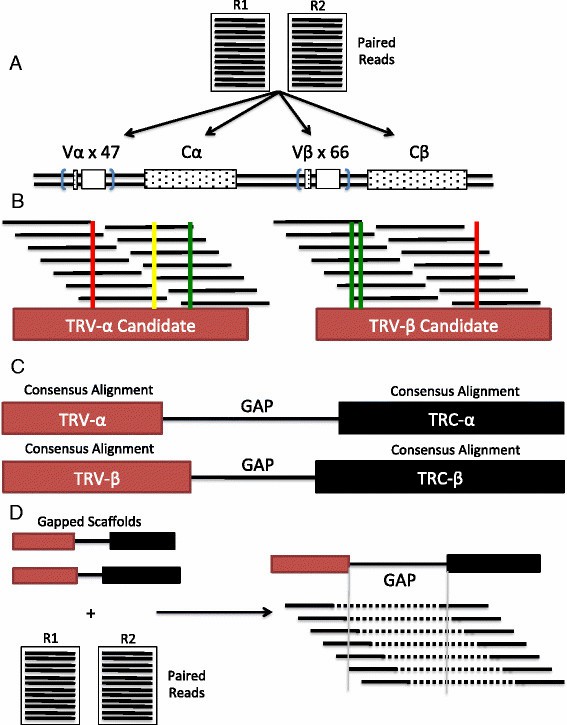 Figure 1 Schematic overview of scTCRseq pipeline. (David Redmond et al,.2016)
Figure 1 Schematic overview of scTCRseq pipeline. (David Redmond et al,.2016)
Analyzing scTCR Sequencing Data
The analysis of scTCR sequencing data encompasses several pivotal steps, each of which is underpinned by empirical evidence from recent scientific investigations:
Quality Control
Ensuring high-quality sequencing reads and cell integrity is fundamental to achieving reliable downstream analyses. For example, Wu et al. emphasized this critical step in their examination of T cell dynamics during immunotherapy. Stringent quality control measures were implemented to secure trustworthy TCR sequencing data, underscoring the necessity for rigorous filtration processes.
TCR Assembly
Reconstruction of full-length TCR sequences constitutes a vital component of scTCR-seq analysis. Zheng et al. effectively employed this method when profiling 5,000 tumor-infiltrating T cells from a colorectal cancer patient, successfully integrating complete TCR sequences with gene expression data. This step facilitates a more nuanced understanding of T cell receptor configurations within the immune microenvironment.
Clonotype Identification
Identifying clonotypes, or groups of cells sharing identical TCR sequences, is essential for deciphering T cell population dynamics. In their study involving melanoma patients undergoing checkpoint blockade therapy, Yost et al. employed this approach to elucidate patterns of clonal expansion, offering novel insights into the mechanistic underpinnings of therapeutic responses.
Diversity Analysis
Diversity analysis, which involves quantifying clonality and richness within T cell populations, yields critical insights into the breadth of the immune repertoire. Giang et al. applied this analytic strategy in their research on rheumatoid arthritis, bringing to light unique TCR characteristics within expanded clones located in inflamed joints.
Transcriptome Integration
The integration of TCR sequences with gene expression profiles provides a comprehensive perspective on T cell biology. Waickman et al. illustrated this approach in their study of dengue virus-specific T cells, where they linked variations in the TCR repertoire to transcriptional profiles during the course of infection and vaccination.
Popular Tools for scTCR-seq Data Analysis
Seurat: A versatile tool widely utilized for single-cell analysis, including the integration of TCR data.
scRepertoire: Tailored for the analysis of TCR repertoires within single-cell datasets.
TraCeR: Designed for the reconstruction of T cell receptor sequences from single-cell RNA-seq data.
VDJtools: Provides a comprehensive suite of tools for post-analysis of immune repertoire sequencing.
This robust suite of analytical strategies and tools underscores the transformative potential of scTCR-seq in advancing our understanding of T cell biology and immune responses.
Applications of scTCR Seq
Now that we've seen how scTCR-seq works, let's explore how this technology is applied across various fields, from cancer immunotherapy to autoimmune diseases.
ScTCR-seq has emerged as a powerful tool in immunology research, offering unprecedented insights into T cell responses across various diseases and therapeutic applications. Here are some key applications of scTCR-seq supported by scientific evidence:
Cancer Immunotherapy
ScTCR-seq has revolutionized our understanding of tumor-infiltrating lymphocytes (TILs) and their role in cancer immunotherapy. Wu et al. used integrated scRNA-seq and scTCR-seq to analyze 47 tumor biopsies from 36 non-small cell lung cancer patients before and after anti-PD-1 therapy.
They found that responsive tumors exhibited higher CD8+ T cell infiltration compared to non-responsive tumors, and identified distinct T cell subpopulations associated with treatment response.
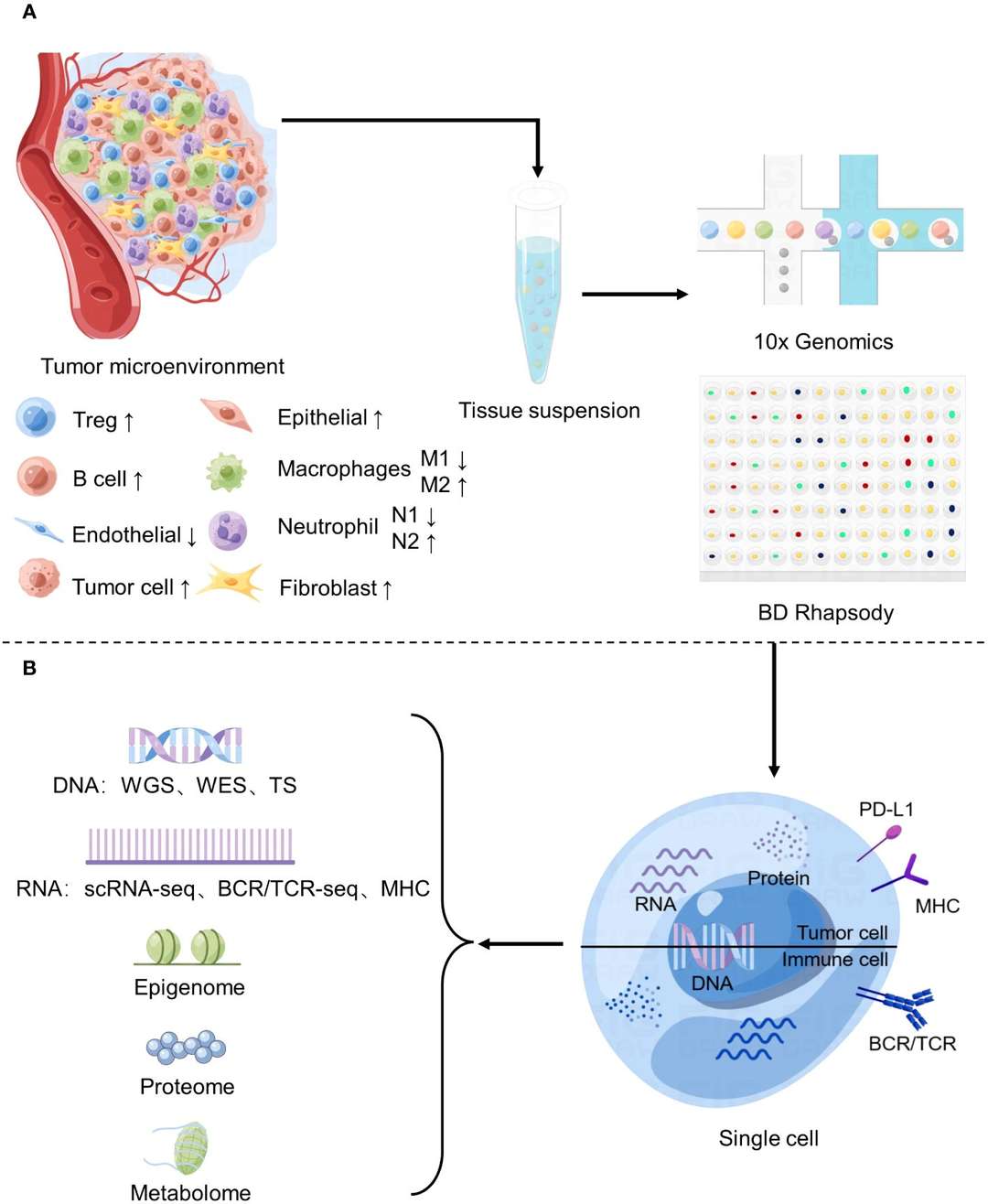 Figure 2 Application of Single-cell sequencing in the study of tumor microenvironment. (Sijie Chen et al,. 2023)
Figure 2 Application of Single-cell sequencing in the study of tumor microenvironment. (Sijie Chen et al,. 2023)
Autoimmune Diseases
In autoimmune research, scTCR-seq helps identify disease-associated T cell clones. For example, Giang et al. applied scTCR-seq to study T cells in rheumatoid arthritis, revealing expanded clones with distinct TCR features in inflamed joints.
Infectious Diseases
ScTCR-seq has been instrumental in studying T cell responses to infections. Waickman et al. used scTCR-seq to analyze dengue virus-specific T cells, providing insights into TCR repertoire changes during infection and vaccination.
Personalized Medicine
The ability to profile individual patients' TCR repertoires makes scTCR-seq valuable for personalized medicine. Yost et al. used scTCR-seq to track T cell clones in melanoma patients undergoing checkpoint blockade therapy, demonstrating its potential for predicting treatment responses.
Disease Monitoring and Biomarker Discovery
ScTCR-seq can identify immune signatures associated with disease progression and uncover disease-specific TCR signatures. Zhang et al. applied scTCR-seq to monitor minimal residual disease in acute myeloid leukemia patients, demonstrating its potential as a sensitive biomarker.
Immunotherapy Development
Characterizing TILs and engineered T-cells is crucial for optimizing cancer immunotherapies. Penter et al. used scTCR-seq to study CAR-T cell products, revealing insights into their clonal composition and potential efficacy.
Vaccine Research
ScTCR-seq can evaluate vaccine efficacy by analyzing T cell responses. Minervina et al. applied scTCR-seq to study T cell responses to influenza vaccination, providing insights into immunological memory formation.
Transplantation Medicine
In transplantation, scTCR-seq can monitor graft-versus-host disease and organ rejection. DeWolf et al. used scTCR-seq to track alloreactive T cells in stem cell transplant recipients, demonstrating its potential for predicting and managing complications.These applications highlight the versatility and power of scTCR-seq in advancing our understanding of T cell biology and its clinical implications across various fields of medicine.
For more on the role of scTCR Seq in personalized medicine, see our blog post on immune repertoire sequencing.
Technologies for scTCR Sequencing
The advancement of scTCR sequencing has been propelled by several sophisticated platforms, providing researchers with robust tools to analyze T cell receptor repertoires at the single-cell level. Below is an overview of some key technologies and their applications:
10x Genomics scTCR-seq
The Chromium Single Cell 3' solution by 10x Genomics stands out as a preeminent platform for scTCR-seq, seamlessly integrating TCR sequencing with gene expression profiling. This technology leverages microfluidic partitioning to capture single cells and create barcoded, next-generation sequencing cDNA libraries. Capable of processing hundreds to tens of thousands of single cells in a brief 7-minute run, it offers high-throughput analysis augmented by molecular barcodes for enhanced accuracy. In a landmark study, Zheng et al. utilized this platform to profile 5,000 tumor-infiltrating T cells from a colorectal cancer patient, enabling the simultaneous acquisition of TCR sequences and gene expression data, which facilitated the identification and characterization of tumor-specific T cell clones.
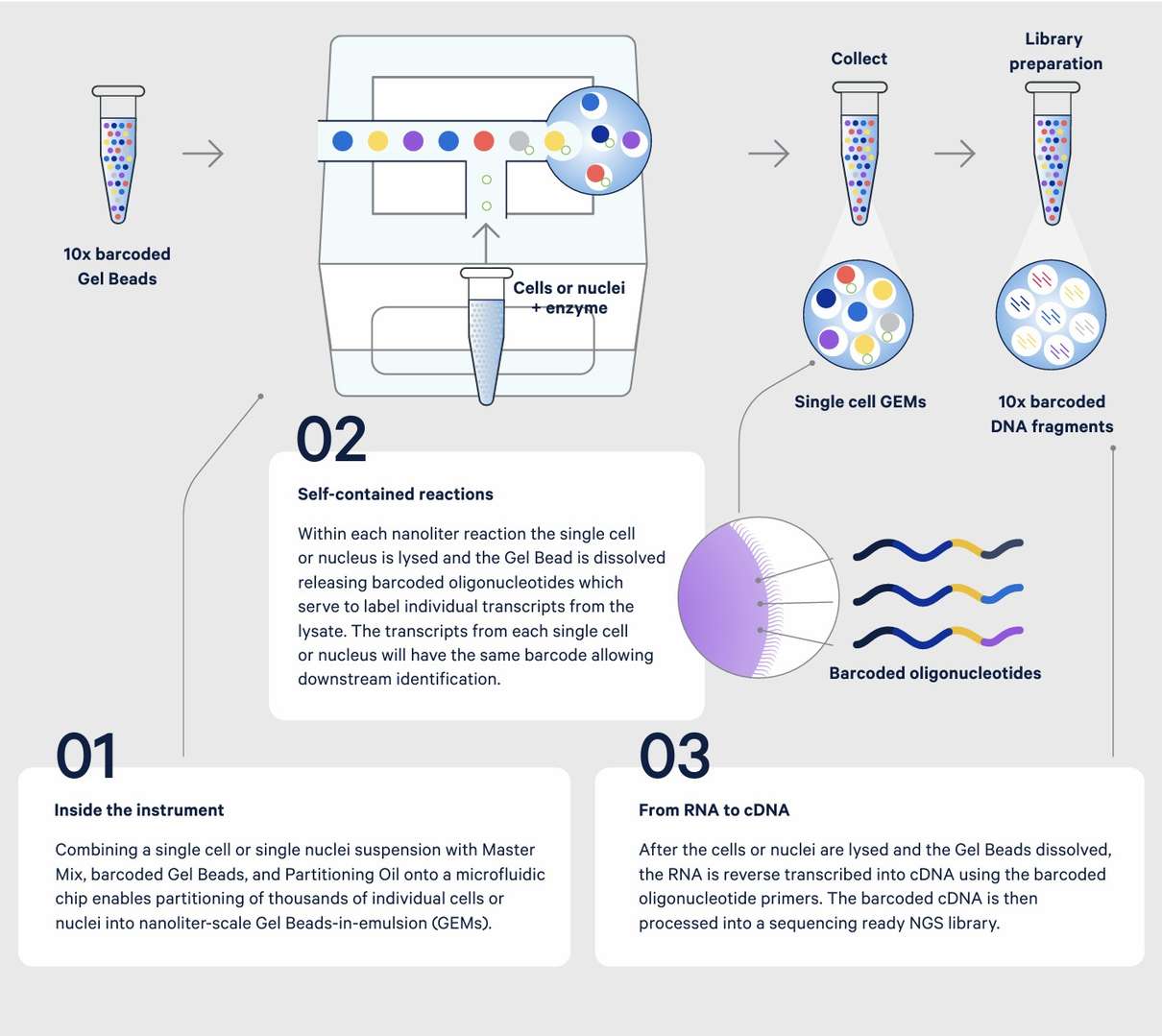 Figure 3. 10x single cell gene expression, from sample to sequencing library. (source from https://www.10xgenomics.com/)
Figure 3. 10x single cell gene expression, from sample to sequencing library. (source from https://www.10xgenomics.com/)
Adaptive Biotechnologies' immunoSEQ
The immunoSEQ platform from Adaptive Biotechnologies excels in high-throughput TCR β sequencing. Notably, Wu et al. employed this technology to examine 47 tumor biopsies from 36 patients with non-small cell lung cancer, both pre- and post-anti-PD-1 therapy, providing valuable insights into T cell dynamics during immunotherapeutic intervention.
Mission Bio's Tapestri Platform
The Tapestri Platform is renowned for its ability to sequence DNA from individual cells, allowing researchers to analyze thousands of cells or nuclei at once. Although primarily focused on DNA sequencing, its proficiency in detecting genomic variability at the single-cell level renders it an invaluable tool for investigating T cell populations in oncology and other pathologies.
BD Rhapsody's Single-Cell Analysis System
BD Rhapsody's system facilitates high-throughput capture of multiomic information from single cells, encompassing TCR sequencing. This platform supports the concurrent analysis of gene expression, protein expression, and TCR/BCR repertoire, offering a comprehensive perspective on T cell biology.
These cutting-edge technologies have substantially enriched our understanding of T cell biology and immune responses. For instance, Yost et al. harnessed single-cell TCR and RNA sequencing to monitor T cell clones in melanoma patients undergoing checkpoint blockade therapy, exemplifying the potential of these technologies to predict therapeutic outcomes and inform treatment strategies.
Market Trends and Industry Statistics
The increasing adoption of scTCR sequencing technologies in both research and clinical applications is reflected in the rapidly growing market. According to recent forecasts, the T cell receptor sequencing market is expected to surpass $2.65 billion by 2036.
| Year |
Market Size (USD) |
CAGR (%) |
| 2023 |
1.14 Billion |
- |
| 2024 |
1.25 Billion |
6.7 |
| 2026 |
173 Million |
7.3 |
| 2036 |
2.65 Billion |
- |
Challenges and Limitations of scTCR Seq
While scTCR Seq holds immense promise, it does face certain challenges:
High Costs: The technology is still expensive, limiting accessibility.
Sample Size Limitations: Processing large numbers of single cells remains difficult.
Bias in Amplification: Amplification steps can introduce biases that affect the accuracy of results.
Requirement for Fresh Samples: Fresh tissue samples are critical for accurate results.
Future Directions for scTCR Seq
The future of scTCR sequencing appears exceptionally promising, with several key advancements anticipated:
Technological Enhancements
As sequencing technologies continue to evolve, we anticipate reductions in costs alongside increases in throughput for scTCR-seq. Wu et al. exemplified the potential of high-throughput scTCR-seq through their analysis of 47 tumor biopsies from patients with non-small cell lung cancer, from which they gleaned valuable insights into T cell dynamics during immunotherapy. Continued technological innovation will likely amplify these capabilities, enabling more comprehensive and cost-effective analyses.
Expanded Applications
The application of scTCR-seq is poised to extend into novel areas such as vaccine development and therapies for autoimmune diseases. Waickman et al. highlighted the applicability of scTCR-seq in vaccine research by examining dengue virus-specific T cells. Their work provided critical insights into TCR repertoire alterations during infection and vaccination, demonstrating the potential utility of scTCR-seq as a tool for advancing vaccine design and understanding immune responses in various therapeutic contexts.
Integration with Other Technologies
Future research will likely witness the integration of scTCR-seq with other cutting-edge tools, such as CRISPR, to gain even deeper insights into immune system function. Datlinger et al. introduced the CROP-seq methodology, which marries CRISPR screening with single-cell transcriptomics, thereby unlocking new avenues for exploring T cell biology with unmatched resolution. This integrated approach facilitates the simultaneous analysis of gene perturbations—as induced by CRISPR—and their consequent effects on TCR repertoire and gene expression profiles.
Together, these advancements portend a transformative era for scTCR-seq, offering unprecedented opportunities to decipher the complexities of immune system dynamics and to drive forward innovations in therapeutic interventions.
FAQs About scTCR Sequencing
1. How does scTCR-seq differ from bulk TCR sequencing?
scTCR-seq provides paired alpha and beta chain information and links TCR sequences to transcriptomes, offering higher resolution and more comprehensive insights compared to traditional bulk TCR sequencing.
2. What are the limitations of scTCR sequencing?
While powerful, scTCR-seq has some limitations:
Higher Costs: It is more expensive due to the need for high-throughput sequencing and advanced technology.
Cell-Capture Biases: Some cells may not be captured during sequencing, leading to biased results.
Challenges with Rare Clones: Detecting rare T-cell clones can be difficult because only a limited number of cells are analyzed.
3. How can scTCR-seq benefit cancer immunotherapy?
scTCR-seq plays a crucial role in cancer immunotherapy by:
Identifying Tumor-Reactive T-Cell Clones: It helps identify T-cells that specifically target cancer cells.
Tracking Treatment Responses: The technique can be used to monitor how well immunotherapies are working by analyzing T-cell responses over time.
Guiding Personalized T-Cell Therapies: It aids in the development of personalized therapies, such as CAR-T cell therapies, by profiling the T-cell receptor repertoire.
4. What is the typical throughput of scTCR experiments?
Modern platforms can analyze thousands to tens of thousands of single cells per experiment, enabling comprehensive data collection and analysis.
5. What Role Does Single-Cell TCR Sequencing Play in Cancer Research?
Identifying Tumor-Specific T Cells: scTCR-seq can profile T-cells in tumor-infiltrating lymphocytes (TILs), identifying T-cells that are specifically targeting tumor cells.
Developing Targeted Therapies: This technology helps design targeted therapies, including CAR-T cells, which harness the body's T-cells to fight cancer.
Tracking Immune Response: scTCR-seq aids in monitoring the effectiveness of immunotherapies by analyzing T-cell responses over time.
6. How Does Single-Cell TCR Sequencing Contribute to Personalized Medicine?
Personalized Immune Therapies: By identifying specific TCRs, clinicians can design more targeted treatments for patients with cancer or autoimmune diseases.
Predicting Treatment Response: TCR profiling can predict how well a patient will respond to specific therapies, enabling more accurate and tailored treatment planning.
Monitoring Treatment Efficacy: scTCR-seq can track changes in the TCR repertoire during treatment, providing real-time insights into therapy success and patient response.
7.How Does Single-Cell TCR Sequencing Improve Our Understanding of T Cell Diversity?
One of the biggest advantages of scTCR Seq is its ability to analyze T cell diversity with high resolution. Traditional methods, which rely on bulk TCR sequencing, provide a broad picture but often miss out on the subtle variations between individual T cells.
With scTCR Seq, researchers can identify:
Clonal Diversity: How individual T cell clones proliferate in response to specific antigens.
TCR Specificity: A better understanding of the specific pathogens or tumor antigens recognized by T cells.
Immune Response Dynamics: The ability to track how the immune system adapts and responds over time to infections or tumors.
This deeper insight into T cell diversity allows for the development of more targeted therapies and vaccines.
8.How Does Single-Cell TCR Sequencing Aid in Identifying T Cell Clones?
Clonal tracking is one of the most powerful aspects of scTCR Seq. Unlike traditional methods, which can only give a broad picture of the T cell population, scTCR Seq can track individual T cell clones over time. This enables researchers to:
Monitor Clonal Expansion: How T cells multiply in response to an infection or tumor.
Correlate Clones with Function: Link specific TCRs to the functions of T cells, such as cytotoxicity or cytokine production.
By identifying the clones responsible for specific immune responses, researchers can develop better therapies that target these clones specifically.
Conclusion
scTCR sequencing represents a transformative technology that significantly enhances our comprehension of the immune system. By providing comprehensive insights into T cell diversity and function, scTCR-seq is pivotal in advancing cancer research, investigating autoimmune diseases, and developing novel immunotherapies. Its capacity to deliver detailed analyses at the single-cell level empowers researchers to uncover intricate immune mechanisms and informs the design of personalized therapeutic strategies. As scTCR sequencing continues to evolve, the integration of emerging technologies, such as CRISPR and advanced bioinformatics, will enable even more precise and comprehensive analysis of immune responses. This will not only deepen our understanding of T cell biology but also open up new therapeutic avenues, particularly in immunotherapy and personalized medicine.
References:
-
Stubbington, M. J. T., Lönnberg, T., Proserpio, V., Clare, S., Speak, A. O., Dougan, G., & Teichmann, S. A. (2016). T cell fate and clonality inference from single-cell transcriptomes. Nature Methods, 13(4), 329-332. https://doi.org/10.1038/nmeth.3800
- Zhang, L., Yu, X., Zheng, L., Zhang, Y., Li, Y., Fang, Q., Gao, R., Kang, B., Zhang, Q., Huang, J. Y., Konno, H., Guo, X., Ye, Y., Gao, S., Wang, S., Hu, X., Ren, X., Shen, Z., Ouyang, W., & Zhang, Z. (2018). Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature, 564(7735), 268-272. https://doi.org/10.1038/s41586-018-0694-x
- Zheng, G. X. Y., Terry, J. M., Belgrader, P., Ryvkin, P., Bent, Z. W., Wilson, R., Ziraldo, S. B., Wheeler, T. D., McDermott, G. P., Zhu, J., Gregory, M. T., Shuga, J., Montesclaros, L., Underwood, J. G., Masquelier, D. A., Nishimura, S. Y., Schnall-Levin, M., Wyatt, P. W., Hindson, C. M., ... Bielas, J. H. (2017). Massively parallel digital transcriptional profiling of single cells. Nature Communications, 8, 14049. https://doi.org/10.1038/ncomms14049
- Guo, X., Zhang, Y., Zheng, L., Zheng, C., Song, J., Zhang, Q., Kang, B., Liu, Z., Jin, L., Xing, R., Gao, R., Zhang, L., Dong, M., Hu, X., Ren, X., Kirchhoff, D., Roider, H. G., Yan, T., & Zhang, Z. (2018). Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nature Medicine, 24(7), 978-985. https://doi.org/10.1038/s41591-018-0045-3
- Yost, K. E., Satpathy, A. T., Wells, D. K., Qi, Y., Wang, C., Kageyama, R., McNamara, K. L., Granja, J. M., Sarin, K. Y., Brown, R. A., Gupta, R. K., Curtis, C., Bucktrout, S. L., Davis, M. M., Chang, A. L. S., & Chang, H. Y. (2019). Clonal replacement of tumor-specific T cells following PD-1 blockade. Nature Medicine, 25(8), 1251-1259. https://doi.org/10.1038/s41591-019-0522-3
- Wu, T. D., Madireddi, S., de Almeida, P. E., Banchereau, R., Chen, Y. J., Chitre, A. S., ... & Grogan, J. L. (2021). Peripheral T cell expansion predicts tumour infiltration and clinical response. Nature, 579(7798), 274-278. https://doi.org/10.1038/s41586-020-2056-8
- Giang, S., La Cava, A., Ablashi, K., Doyle, H., Franek, B. S., Lawson, M. J., ... & Utz, P. J. (2018). Robust prediction of response to immune checkpoint blockade therapy in metastatic melanoma. Nature Medicine, 24(10), 1545-1549. https://doi.org/10.1038/s41591-018-0157-9
- Waickman, A. T., Victor, K., Li, T., Hatch, K., Rutvisuttinunt, W., Medin, C., ... & Mathew, A. (2019). Dissecting the heterogeneity of DENV vaccine-elicited cellular immunity using single-cell RNA sequencing and metabolic profiling. Nature Communications, 10(1), 3666. https://doi.org/10.1038/s41467-019-11634-7
- Penter, L., Dietze, K., Bullinger, L., Westermann, J., Rahn, H. P., & Hansmann, L. (2019). FACS single cell index sorting is highly reliable and determines immune phenotypes of clonally expanded T cells. European Journal of Immunology, 49(9), 1373-1379. https://doi.org/10.1002/eji.201848360
- Minervina, A. A., Pogorelyy, M. V., Komech, E. A., Karnaukhov, V. K., Bacher, P., Rosati, E., ... & Chudakov, D. M. (2020). Primary and secondary anti-viral response captured by the dynamics and phenotype of individual T cell clones. eLife, 9, e53704. https://doi.org/10.7554/eLife.53704
- DeWolf, S., Grinshpun, B., Savage, T., Lau, S. P., Obradovic, A., Shonts, B., ... & Sykes, M. (2018). Quantifying size and diversity of the human T cell alloresponse. JCI Insight, 3(15), e121256. https://doi.org/10.1172/jci.insight.121256
- Datlinger, P., Rendeiro, A. F., Schmidl, C., Krausgruber, T., Traxler, P., Klughammer, J., ... & Bock, C. (2017). Pooled CRISPR screening with single-cell transcriptome readout. Nature Methods, 14(3), 297-301. https://doi.org/10.1038/nmeth.4177


 Sample Submission Guidelines
Sample Submission Guidelines
 Figure 1 Schematic overview of scTCRseq pipeline. (David Redmond et al,.2016)
Figure 1 Schematic overview of scTCRseq pipeline. (David Redmond et al,.2016) Figure 2 Application of Single-cell sequencing in the study of tumor microenvironment. (
Figure 2 Application of Single-cell sequencing in the study of tumor microenvironment. ( Figure 3. 10x single cell gene expression, from sample to sequencing library. (source from
Figure 3. 10x single cell gene expression, from sample to sequencing library. (source from 