Throughout human history, the immune system has represented our most formidable line of defense against disease. Central to this defense are T cells and B cells, which function as the primary components of adaptive immunity. These cells are equipped with specialized receptors on their surfaces-T-cell receptors (TCRs) and B-cell receptors (BCRs)-which enable them to recognize and respond to pathogens. Through immune repertoire sequencing, these crucial receptors can be thoroughly examined, providing valuable insights into their role in maintaining health and combating disease. By profiling the diversity and adaptability of TCRs and BCRs, this technology offers a comprehensive understanding of immune system strategies, shedding light on immune responses, disease resistance mechanisms, and potential avenues for therapeutic intervention.
What are TCRs and BCRs?
TCRs and BCRs constitute critical molecular components of the immune system, predominantly situated on the surfaces of T cells and B cells, respectively. These receptors play specialized roles in recognizing and responding to specific antigens, thus forming a cornerstone of the immune response.
Structure and Composition of TCRs
TCRs are composed of two distinct polypeptide chains, typically an alpha (α) and a beta (β) chain. However, in certain subsets of T cells, these chains may alternatively consist of gamma (γ) and delta (δ) variants. Each chain is characterized by the presence of a variable (V) region and a constant © region, with the variable region being chiefly implicated in antigen recognition. This structural variability is crucial, allowing TCRs to bind a vast array of antigens, a fundamental aspect of adaptive immunity.
Structure and Composition of BCRs
In contrast, BCRs are membrane-bound immunoglobulins (mIg) expressed on the surface of B cells. These receptors can exist in five isotypic forms: IgM, IgD, IgG, IgA, and IgE. Each BCR is structured with two identical heavy chains and two identical light chains, forming a Y-shaped configuration. This structural design equips BCRs to effectively recognize a diverse range of antigens, playing an indispensable role in humoral immunity.
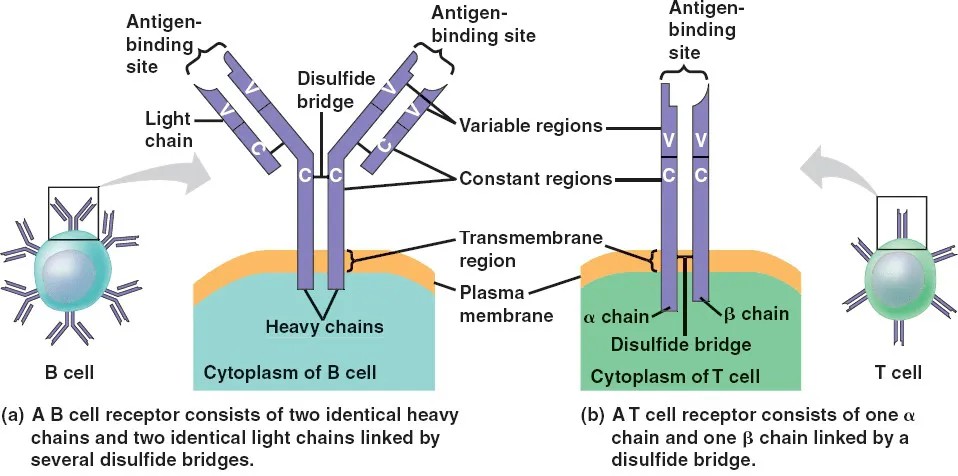 Figure 1.Structure and Composition of TCRs and BCRs
Figure 1.Structure and Composition of TCRs and BCRs
The Functional Roles and Mechanisms of TCR and BCR in the Immune System
TCR Types and Their Distinct Roles
There are two major TCR classes: αβ TCRs and γδ TCRs. The majority of T cells express αβ TCRs, which predominantly recognize endogenous antigens presented by MHC class I molecules, including peptide fragments derived from viral or tumor-associated proteins. Conversely, γδ TCRs are more commonly found in specific tissues, such as the gastrointestinal tract and skin. These γδ TCRs exhibit unique capabilities by recognizing non-peptidic antigens, such as lipids and metabolic byproducts, thus playing specialized roles in certain immune responses.
BCR Function and Activation
In contrast to TCRs, BCRs directly recognize and bind free antigens independently of MHC molecule involvement. BCR specificity is highly refined; each B cell expresses a unique BCR that can identify a specific antigen. Upon binding its specific antigen, the B cell undergoes activation, potentially differentiating into either a plasma cell or a memory B cell. Activated plasma cells produce large quantities of antibodies, which share the antigen-binding site with the original BCR. These antibodies perform critical functions, including antigen neutralization and enhancement of antigen clearance from the body, thus supporting adaptive immune defense mechanisms.
Experimental Procedure
Principle of RNA Immuno-Repertoire Library Construction: molecular barcodes and 5' RACE
The construction of RNA-based immuno-repertoire libraries utilizing molecular barcodes in conjunction with 5' Rapid Amplification of cDNA Ends (5' RACE) permits high-resolution profiling of immune receptor diversity. This methodology proves particularly effective for capturing the native transcript abundance of immunoglobulin (Ig) and TCR RNA, thereby revealing insights into both sequence diversity and expression levels.
5' RACE for Comprehensive Receptor Profiling
The 5' RACE technique is extensively employed to capture the full-length variable (V) regions of Ig and TCR transcripts from their 5' termini. This methodology initiates with reverse transcription, wherein a specific primer anneals to the constant © region of the transcript, aiding the synthesis of complementary DNA (cDNA). Subsequent procedural steps involve capturing and amplifying the 5' end, enabling researchers to retrieve the complete V-region, which is pivotal for comprehending the antigen-binding diversity of immune receptors. This technique is particularly advantageous as it captures the transcript's natural inception point, ensuring a more accurate representation of the VDJ region's architecture.
Molecular barcodes for Reducing Amplification Bias
Incorporating molecular barcodes-short, random nucleotide sequences appended to each RNA molecule prior to amplification-plays a vital role in controlling amplification bias and facilitating transcript abundance quantification. Molecular barcodes are integrated during the initial reverse transcription phase, with each RNA molecule being tagged with a distinctive sequence. During subsequent amplification and sequencing processes, these molecular barcodes enable the differentiation of original RNA molecules from PCR duplicates, allowing for an accurate assessment of transcript abundance and reducing amplification artifacts. This step is critical for achieving quantitative insights into immune repertoire diversity, permitting reliable normalization across disparate transcripts.
Combined Use in High-Throughput Sequencing
Together, the strategies of 5' RACE and molecular barcodes foster a high-throughput, quantitative approach to RNA immuno-repertoire profiling. While 5' RACE ensures comprehensive capture of the immune receptor's V-region, molecular barcodes improve accuracy in quantifying the original RNA molecules. When integrated with next-generation sequencing, these methodologies facilitate an in-depth characterization of immune repertoires, providing essential data for research in immunology, vaccine development, and immune-related disease exploration.
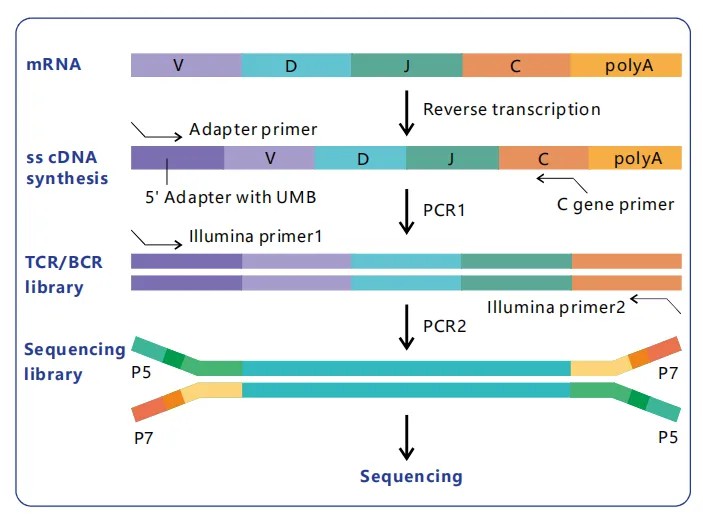 Figure 2. Principle of RNA Immuno-Repertoire Library Construction
Figure 2. Principle of RNA Immuno-Repertoire Library Construction
Advantages and Limitations of RNA-Based Immuno-Repertoire Library Construction
Increased Sensitivity: RNA-based approaches offer enhanced sensitivity due to the presence of multiple mRNA copies per cell, which facilitates amplification and molecular barcodes integration.
Error Correction through molecular barcodes: The introduction of molecular barcodes technology aids in reconstructing the pre-PCR amplification state, allowing for the correction of errors arising from PCR and sequencing processes.
Comprehensive Sequence Capture with 5' RACE: The 5' rapid amplification of cDNA ends (5' RACE) method enables amplification of full-length immuno-repertoire gene sequences. This technique not only reveals novel exon genes and mutation types but also adapts to various sequencing read-length formats, thereby permitting the acquisition of full-length sequences of the CDR3 region or complete TCR/BCR sequences.
Sample Quality Requirement: This method necessitates high-quality samples to achieve accurate and reproducible results.
Principle of DNA-Based Immuno-Repertoire Library Construction: Multiplex PCR Technology
The construction of DNA-based immuno-repertoire libraries is predicated on the utilization of multiplex PCR technology, a robust methodology adept at simultaneously amplifying multiple gene segments within a singular reaction framework. This methodological approach is especially advantageous in the exploration of immune repertoires, facilitating the exhaustive capture of Ig and TCR gene segments. Such segments are characterized by considerable diversity, a consequence of recombination and mutation events.
In the construction of immuno-repertoire libraries, multiplex PCR leverages meticulously crafted primer pairs, which are strategically designed to target the variable, diversity, and joining (VDJ) regions of Ig or TCR genes. These primers must exhibit high specificity to engender the amplification of a comprehensive array of unique VDJ combinations while concurrently curtailing off-target amplification. Through the precise design of primers, this technique succeeds in encapsulating a wide spectrum of immune receptor diversity within a given sample, a critical factor for applications such as immune profiling, disease research, and therapeutic development.
Furthermore, during the amplification process, barcoding sequences are frequently incorporated into the PCR products. These barcodes facilitate the differentiation of individual samples in subsequent high-throughput sequencing phases, thereby permitting accurate tracking and comparative analysis across a multitude of libraries. This step proves particularly beneficial in large-scale studies by enabling the parallel processing of numerous samples. Consequently, it significantly reduces both temporal and financial investment while concurrently augmenting the depth of analytical inquiry.
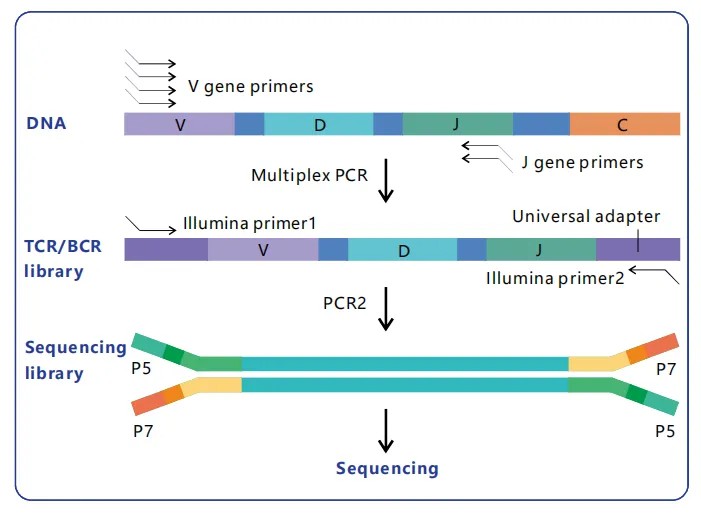 Figure 3. Principle of DNA Immuno-Repertoire Library Construction
Figure 3. Principle of DNA Immuno-Repertoire Library Construction
Advantages and Limitations of DNA-Based Immuno-Repertoire Library Construction
- Suitability for Degraded Samples: DNA-based methods are well-suited for samples where RNA quality cannot be reliably maintained, such as degraded RNA samples or formalin-fixed, paraffin-embedded (FFPE) specimens.
- Quantitative Analysis Capability: DNA-based approaches enable relatively quantitative analysis of lymphocyte populations, including T and B cells, since each copy of genomic DNA (gDNA) correlates directly to a single cell, providing a one-to-one basis for cell quantification.
- Targeted Amplification of the CDR3 Region: This method primarily amplifies the complementarity-determining region 3 (CDR3), a critical segment of immunoglobulin and T-cell receptor genes responsible for antigen binding and specificity.
- Lower Sample Quality Requirements: DNA-based immuno-repertoire construction requires lower sample quality compared to RNA-based methods, facilitating easier handling and analysis.
You may interested in
Learn More
Applications of TCR/BCR Research Precision Medicine
The analysis of tumor-specific antigens on patient-derived cancer cells allows for the development of personalized TCR-T (T-cell receptor-transduced T-cell) therapies. For example, TCR-T cell therapy targeting melanoma has emerged as a promising approach in oncology.
- Title: Functional Analysis of Peripheral and Intratumoral Neoantigen-Specific TCRs Identified in a Patient with Melanoma
- Journal: Journal for ImmunoTherapy of Cancer
- Impact Factor: 10.3
- Publication Date: September 2021
Summary
A case study of metastatic malignant melanoma was conducted to characterize neoantigen-specific T-cell responses within the framework of immune checkpoint regulation. Researchers employed immunopeptidomics and computational prediction to identify neoantigens, followed by TCR sequencing to isolate TCRs specific to these neoantigens from the patient's immune repertoire.
Subsequent in vitro multiparametric analyses-including functional affinity assessments, multiplex cytokine secretion assays, and cross-reactivity screening-were utilized to evaluate the antitumor efficacy of these TCRs. Selected mutated peptide ligands demonstrated immunogenic potential, exhibiting binding affinities comparable to human leukocyte antigen (HLA) complexes. These peptide ligands also displayed differential binding characteristics relative to their wild-type counterparts in molecular dynamics simulations.
Interestingly, TCRs recognizing these neoantigens exhibited varied functional and frequency profiles. TCRs with comparatively lower functional affinity displayed robust antitumor immune responses in vivo, a finding that underscores the potential effectiveness of these TCRs despite their moderate affinity. Additionally, these TCRs were detected at high frequencies, particularly in tumor tissue, lymph nodes, and various blood samples, demonstrating prolonged persistence. This persistence aligns with a reduced activation pattern that follows primary in vitro stimulation, highlighting the potential for long-term antitumor efficacy in the tumor microenvironment.
Immunotherapy Applications
Utilizing the specificity of TCRs and BCRs has enabled the design of innovative bispecific antibodies, such as Blinatumomab, targeting specific leukemia subtypes and aiding in the assessment of therapeutic efficacy for various malignancies.
- Title: Neoadjuvant Tislelizumab Plus Stereotactic Body Radiotherapy and Adjuvant Tislelizumab in Early-Stage Resectable Hepatocellular Carcinoma: The Notable-HCC Phase Ib Trial
- Journal: Nature Communications
- Impact Factor: 16.6
- Publication Date: April 2024
Study Design
This investigation, a single-center, open-label, phase Ib clinical trial (NCT05185531), enrolled 20 patients diagnosed with Barcelona Clinic Liver Cancer (BCLC) stage 0-A hepatocellular carcinoma (HCC). Participants received three sessions of stereotactic body radiotherapy (SBRT) in combination with two cycles of neoadjuvant PD-1 antibody therapy (tislelizumab). Tumor tissue and peripheral blood samples were collected before and after surgical intervention for pathological assessment, immunohistochemistry, RNA sequencing, and TCR sequencing.
Primary Endpoints
Key endpoints included surgical delay, radiologic and pathologic tumor response, and safety assessment. Preliminary results indicated favorable safety and antitumor efficacy for the combination of neoadjuvant PD-1 inhibition and SBRT in patients with early-stage resectable HCC. Additionally, TCR sequencing proved instrumental in evaluating immune activation following neoadjuvant therapy and elucidating its mechanisms.
Evaluating Immunotherapy Efficacy through TCR Repertoire Analysis
The efficacy of neoadjuvant PD-1 antibody immunotherapy was assessed by analyzing changes in the TCR repertoire pre- and post-treatment. Post-treatment analysis revealed a substantial increase in novel TCR clones, suggesting a robust immunotherapeutic response. TCR sequencing identified a significant presence of new, high-frequency TCR clones, further supporting the hypothesis that neoadjuvant treatment promotes antitumor effects through the expansion and turnover of tumor-specific T-cell populations.
Therapeutic Strategies for Autoimmune Diseases
Advancements in the analysis of aberrant T-cell receptor (TCR) expression patterns in autoimmune diseases offer potential pathways for the development of TCR-targeting therapeutic agents. Such therapeutics are designed to modulate TCR activity, thereby attenuating autoimmune responses. For instance, biological agents tailored for conditions such as rheumatoid arthritis exemplify this approach.
- Title: Analysis of the B-Cell Receptor Repertoire in Six Immune-Mediated Diseases
- Journal: Nature
- Impact Factor: 42.778
- Publication Date: September 2019
Study Overview
This study undertook a comprehensive analysis of B-cell receptor (BCR) repertoires across six autoimmune diseases, specifically examining peripheral blood mononuclear cells (PBMCs) from 209 patients with systemic lupus erythematosus (SLE), anti-neutrophil cytoplasmic antibody (ANCA)-associated vasculitis, Crohn's disease, Behçet's disease, eosinophilic granulomatosis with polyangiitis (EGPA), and IgA vasculitis. Additionally, PBMC samples from 19 healthy controls were analyzed. The BCRs within these samples were sequenced using the Illumina MiSeq platform with paired-end 300 bp reads (PE 300).
Methodology
To ensure high-quality sequencing of the BCRs, the study implemented a multiplex PCR approach utilizing primers designed for the V(D)J regions, complemented by universal primers targeting constant (C) regions. This strategy enabled robust amplification and the subsequent construction of a comprehensive BCR sequencing library. The data obtained facilitated a multi-layered analytical framework, which examined:
Influence of Disease Status and Age: Associations between BCR characteristics and both disease state and patient age were evaluated.
Antibody Subtype Distribution: Variation in antibody isotypes was examined across different disease conditions.
IGHV Gene Usage: Frequency of immunoglobulin heavy variable (IGHV) gene usage was assessed to identify any disease-specific patterns.
CDR3 Length and Clonotype Diversity: Comparative analyses of the complementary-determining region 3 (CDR3) length and the diversity of BCR clonotypes were conducted to elucidate disease-specific B-cell clonal expansion.
Class Switching: Patterns of isotype switching were investigated to understand immune adaptation mechanisms within disease contexts.
Longitudinal BCR Variability in Response to Treatment: For a subset of patients undergoing specific treatments, the evolution of BCR repertoires was monitored to discern treatment-induced changes.
Conclusions and Implications
This study provides an extensive characterization of BCR repertoires in autoimmune diseases, revealing intricate structural features and variations associated with disease states. These insights contribute valuable knowledge to the pathogenic mechanisms underlying autoimmune diseases and pave the way for more targeted therapeutic strategies.

Conclusion
During pathogenic invasion, the host immune system mobilizes a variety of immune cells, each characterized by distinct transcriptional profiles. Antigens are recognized through the B-cell receptor (BCR) on B cells and the T-cell receptor (TCR) on T cells, with subsets of these cells undergoing clonal expansion, forming populations that express identical BCR or TCR sequences. This phenomenon elucidates the multidimensional heterogeneity of the immune response when encountering pathogens. By analyzing the sequence diversity of BCRs and TCRs, insights can be gained into the clonal distribution within the immune microenvironment, advancing understanding of the mechanisms through which the organism mounts a defense against pathogen invasion.
TCR sequencing technology demonstrates significant potential within immunotherapeutic research. It not only facilitates progress in the field of personalized medicine but also holds considerable importance in the discovery and optimization of novel immunotherapies.
References:
-
Bräunlein E, Lupoli G, Füchsl F, et al. Functional analysis of peripheral and intratumoral neoantigen-specific TCRs identified in a patient with melanoma. J Immunother Cancer. 2021;9(9):e002754.
- Bashford-Rogers RJM, Bergamaschi L, McKinney EF, et al. Analysis of the B cell receptor repertoire in six immune-mediated diseases. Nature. 2019;574(7776):122-126.
- Li, Z., Liu, J., Zhang, B. et al. Neoadjuvant tislelizumab plus stereotactic body radiotherapy and adjuvant tislelizumab in early-stage resectable hepatocellular carcinoma: the Notable-HCC phase 1b trial. Nat Commun 15, 3260 (2024).


 Sample Submission Guidelines
Sample Submission Guidelines
 Figure 1.Structure and Composition of TCRs and BCRs
Figure 1.Structure and Composition of TCRs and BCRs Figure 2. Principle of RNA Immuno-Repertoire Library Construction
Figure 2. Principle of RNA Immuno-Repertoire Library Construction Figure 3. Principle of DNA Immuno-Repertoire Library Construction
Figure 3. Principle of DNA Immuno-Repertoire Library Construction

