Case Study
Sanger seduencing Implementation in Clinical Ill Patients for Bacterial and
Fungal Patogens Identification
The study presented examines the application of PCR and Sanger sequencing for the rapid diagnosis of bacterial and fungal pathogens in clinical settings [4].
This approach focuses on three primary aspects:
1. Identification of pathogens in critically ill patients through the detection of 16S and 18S/eEF1 gene sequences.
2. Evaluation of the correlation between Sanger sequencing results and conventional biomarkers such as procalcitonin, C-reactive protein, erythrocyte sedimentation rate, and neutrophil-to-lymphocyte ratio.
3. Comparison of molecular diagnostic methods with traditional culture techniques to analyze discrepancies in diagnostic outcomes.
Methodology
1. PCR and Gene Markers
The study involved 30 patients diagnosed with infectious diseases upon admission. Samples were collected, including whole blood, cerebrospinal fluid, bronchoalveolar lavage fluid, and ascitic fluid.
DNA Extraction and PCR Amplification:
PCR was performed to screen for the presence of pathogenic microbial DNA fragments. The specific gene markers used were:
16S rDNA genes (V3-V4 region): Expected product length 400 bp
- Forward primer: CCGTCAATTCCTTTGAGTT
- Reverse primer: CAGCAGCCGCGCTAATAC
eEF1: Expected product length 600 bp
- Forward primer: GAYTTCATCAAGAACATGA
- Reverse primer: GACGTTGAADCCRACRTTG
18S rDNA: Expected product length 150 bp
- Forward primer: GATCACACCGCCCGTC
- Reverse primer: TGATCCTTCTGCAGGTTCA
2. Sanger Sequencing
Amplified products were further analyzed using Sanger sequencing. Sequencing reactions utilized the Big Dye Terminator technique (Thermo Fisher Scientific) and were detected using the 3500 Genetic Analyzer (Applied Biosystems).
3. Data Analysis
Sequence data were analyzed using Geneious Prime v2019.2.3 (https://www.geneious.com/) and compared against the GenBank database (https://www.ncbi.nlm.nih.gov/).
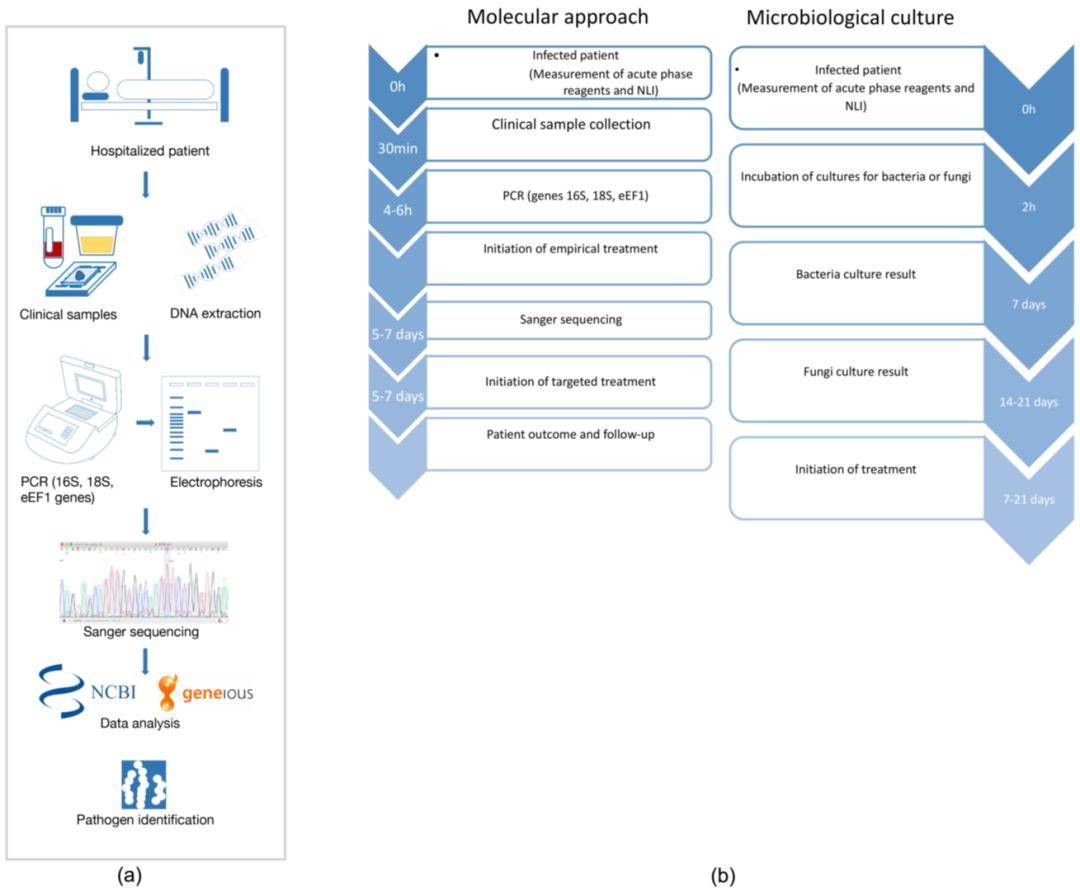 Figure 5. Study Workflow for Identifying Bacterial and Fungal Pathogens Using Sanger Sequencing
Figure 5. Study Workflow for Identifying Bacterial and Fungal Pathogens Using Sanger Sequencing
Results
1. Quality Assessment of Sanger Sequencing
Quality assessment of Sanger sequencing data involved semi-quantitative PCR of the 16S and 18S/eEF1 gene markers. Sequencing quality was evaluated based on base call accuracy, peak height distribution, secondary peak appearance, and overall sequence quality (Figure 1a). Statistical analysis revealed no significant differences in read quality between genes (Figure 1e).
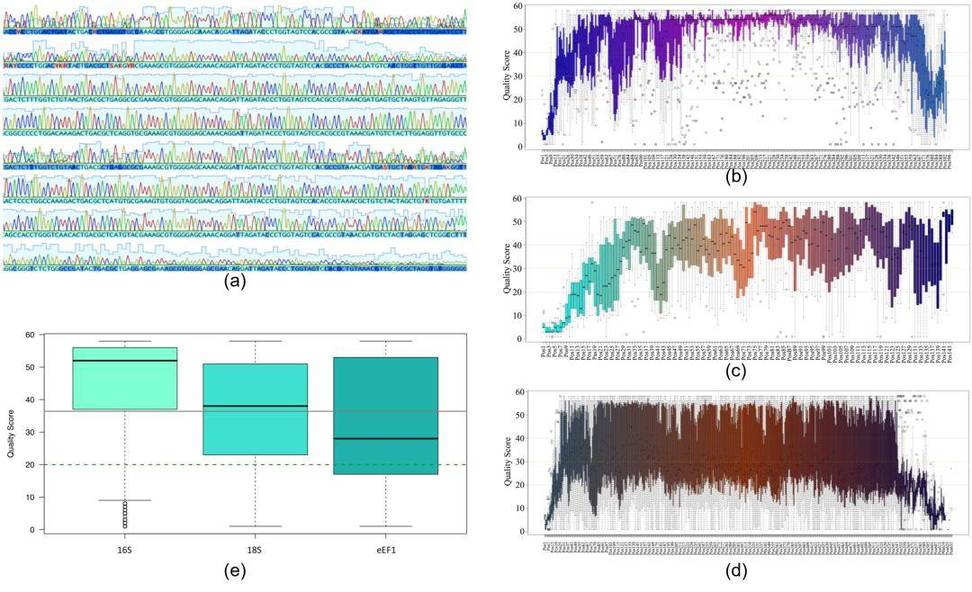 Figure 1: Quality analysis of Sanger sequencing reads for all genomic markers used in the study. (a) Representative electropherogram; (b) Nucleotide quality analysis for 16S; (c) 18S; (d) eEF1 sequences; (e) Quality score distribution for nucleotide positions in 16S, 18S, and eEF1 sequences.
Figure 1: Quality analysis of Sanger sequencing reads for all genomic markers used in the study. (a) Representative electropherogram; (b) Nucleotide quality analysis for 16S; (c) 18S; (d) eEF1 sequences; (e) Quality score distribution for nucleotide positions in 16S, 18S, and eEF1 sequences.
2. Clinical Gene Detection Results
Sequence analysis of bacterial 16S and fungal 18S/eEF1 genes from clinical samples revealed:
46.6% of samples were positive for fungal infections.
23.3% of samples were positive for bacterial infections.
23.3% of samples were positive for both bacterial and fungal infections.
6.7% of samples were negative for any detected pathogens.
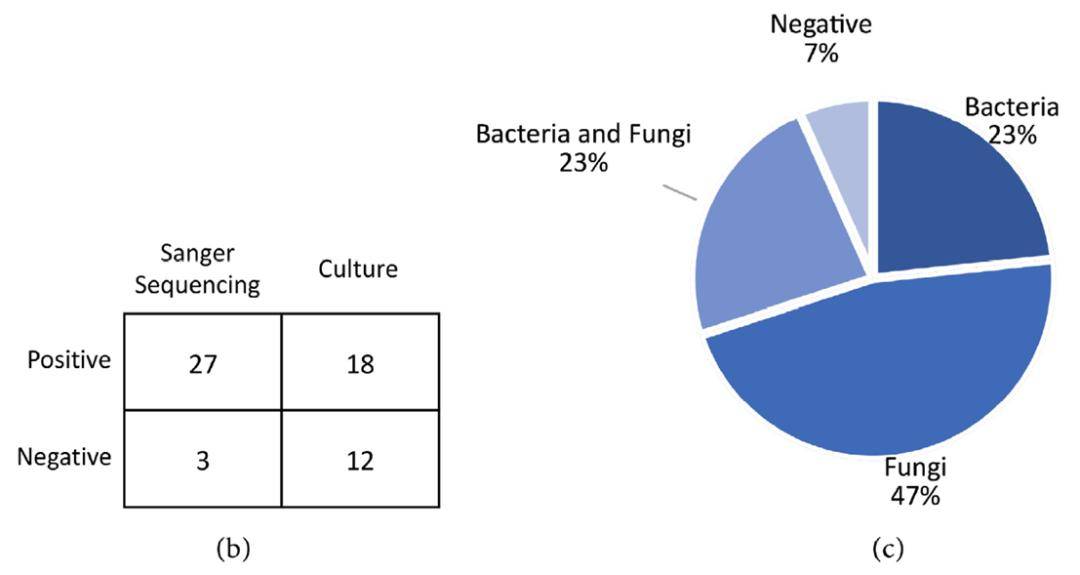 Figure 2: (b) Comparison of Sanger sequencing and culture results; (c) Classification of pathogens identified in clinical samples using PCR+Sanger sequencing.
Figure 2: (b) Comparison of Sanger sequencing and culture results; (c) Classification of pathogens identified in clinical samples using PCR+Sanger sequencing.
3. Comparison: Sanger Sequencing vs. Traditional Culture Methods
Sanger sequencing demonstrated a lower correlation with microbial culture results; many cultures failed to grow and did not align with sequencing findings. Contrarily, Sanger sequencing results directly corresponded with the clinical symptoms of the patients. A statistically significant difference was observed between Sanger sequencing and culture results (p = 9.47×10−8). Out of 30 cases, 28 (93.33%) infections were identified using Sanger sequencing, compared to 18 (60%) identified via culture methods (Figure 2b).
Conclusion
The use of PCR in conjunction with Sanger sequencing for the analysis of 16S and 18S/eEF1 gene markers significantly optimizes the time required to obtain results for bacterial or fungal infections. Sanger sequencing provides a reliable method for pathogen identification, which aids in preventing disease progression and enables the formulation of more targeted therapeutic strategies, ultimately improving clinical outcomes.
Future Directions in Pathogen Identification
Integration with Emerging Technologies
The future of pathogen identification lies in the integration of Sanger sequencing with emerging technologies. Advances in microfluidics, automated sample processing, and real-time sequencing technologies promise to enhance the efficiency and accessibility of molecular diagnostics. Combining Sanger sequencing with these innovations could lead to even faster and more accurate pathogen identification in clinical settings.
Development of Novel Genetic Markers
Ongoing research aims to identify novel genetic markers that improve pathogen detection and differentiation. The development of new markers and sequencing techniques will further enhance the resolution of pathogen identification and expand the applicability of Sanger sequencing to a broader range of infectious agents.
Personalized Medicine and Targeted Therapies
The advancement of pathogen identification techniques is poised to significantly bolster the field of personalized medicine. Specifically, the precise identification capabilities of Sanger sequencing facilitate the development of targeted therapies that are customized to combat specific infections. This precision medicine approach not only holds the potential to enhance treatment efficacy but also aims to mitigate the ongoing issue of antibiotic resistance by reducing the dependence on broad-spectrum antibiotics.
Conclusion
Sanger sequencing remains a valuable tool for the rapid and accurate identification of bacterial and fungal pathogens. Its ability to provide detailed genetic information complements traditional methods and offers significant advantages in clinical diagnostics. As advancements in molecular biology and technology continue to evolve, Sanger sequencing, in conjunction with other emerging techniques, will play a pivotal role in enhancing pathogen detection and guiding effective treatment strategies.
For comprehensive and reliable pathogen identification services, CD Genomics remains at the forefront of integrating advanced molecular techniques to support research applications.
References
- Drancourt, M., Bollet, C., Carlioz, A., et al. (2000) 16S Ribosomal DNA Sequence Analysis of a Large Collection of Environmental and Clinical Unidentifiable Bacterial Isolates. Journal of Clinical Microbiology, 38, 3623-3630.
- Kidd, S., Halliday, C., Alexiou, H. and Ellis, D. (2016) Descriptions of Medical Fungi. 3rd Edition, CutCut Digital, Paris.
- Balajee, S.A., Houbraken, J., Verweij, P.E., et al. (2007) Aspergillus Species Identification in the Clinical Setting. Studies in Mycology, 59, 39-46.
- Fong-Flores, V. , Murillo-Gallo, A. , Contreras-Ojeda, L. and Barraza, A. (2022) Sanger Sequencing Implementation in Clinically Ill Patients for Bacterial and Fungal Pathogens Identification. Advances in Infectious Diseases, 12, 363-382.


 Figure 5. Study Workflow for Identifying Bacterial and Fungal Pathogens Using Sanger Sequencing
Figure 5. Study Workflow for Identifying Bacterial and Fungal Pathogens Using Sanger Sequencing Figure 1: Quality analysis of Sanger sequencing reads for all genomic markers used in the study. (a) Representative electropherogram; (b) Nucleotide quality analysis for 16S; (c) 18S; (d) eEF1 sequences; (e) Quality score distribution for nucleotide positions in 16S, 18S, and eEF1 sequences.
Figure 1: Quality analysis of Sanger sequencing reads for all genomic markers used in the study. (a) Representative electropherogram; (b) Nucleotide quality analysis for 16S; (c) 18S; (d) eEF1 sequences; (e) Quality score distribution for nucleotide positions in 16S, 18S, and eEF1 sequences. Figure 2: (b) Comparison of Sanger sequencing and culture results; (c) Classification of pathogens identified in clinical samples using PCR+Sanger sequencing.
Figure 2: (b) Comparison of Sanger sequencing and culture results; (c) Classification of pathogens identified in clinical samples using PCR+Sanger sequencing.