Structure of Eubacteria: Unveiling the Intricacies of Bacterial Architecture
The architecture of Eubacteria exhibits a sophisticated organization finely calibrated to facilitate their multifaceted metabolic functions, ecological engagements, and adaptive behaviors in response to environmental fluctuations. Through dissecting the intricacies of bacterial structural arrangements, we illuminate the underlying principles dictating bacterial physiology, evolutionary dynamics, and pathogenicity, thereby establishing a bedrock for pioneering methodologies in microbiological studies, biotechnological applications, and antimicrobial investigations.
Cell Wall Composition:
The cell wall stands as a pivotal structural element within Eubacteria, furnishing essential attributes of form, robustness, and safeguarding against osmotic perturbations. Predominantly fashioned from peptidoglycan in most Eubacterial species, this intricate polymer comprises elongated sequences of alternating amino sugars, namely N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM), interlinked by abbreviated peptide chains. This distinctive constitution imparts upon the cell wall its characteristic durability and resilience against mechanical strains, empowering bacteria to uphold their morphology and structural integrity amidst diverse environmental milieus.
Capsule and Glycocalyx:
Certain Eubacteria extend their structural complexity beyond the cell wall by enveloping themselves in an external layer termed the capsule, comprised of polysaccharides or polypeptides. This capsule assumes multifarious roles, acting as a shield against desiccation, phagocytosis, and immune assaults, while also aiding in surface adhesion and biofilm assembly. Furthermore, specific bacterial strains secrete a mucilaginous layer of polysaccharides, termed glycocalyx, which augments cellular adhesion and biofilm development, thereby facilitating bacterial colonization and enduring presence within distinct ecological niches.
Plasma Membrane and Cytoplasm:
Situated beneath the cell wall resides the plasma membrane, a phospholipid bilayer interwoven with proteins, orchestrating the ingress and egress of molecules across the cellular boundary. This pivotal membrane assumes indispensability in myriad cellular functions, including nutrient acquisition, energy generation, signal transduction, and intracellular transport phenomena. Meanwhile, within the cytoplasm, a dense aqueous milieu harboring enzymes, ribosomes, nucleic acids, and assorted cellular constituents hosts a tapestry of metabolic reactions, underpinning bacterial proliferation, propagation, and adaptability within ever-changing surroundings.
Nucleoid and Plasmids:
In contrast to eukaryotic counterparts, Eubacteria are devoid of a discernible nucleus, housing their genetic material within a distinct locale termed the nucleoid. This nucleoid encompasses a solitary circular chromosome, housing pivotal genetic instructions pivotal for cellular operations and hereditary attributes. Moreover, certain bacterial strains harbor supplementary extrachromosomal DNA entities known as plasmids, capable of autonomous replication and endowing adaptive benefits such as antibiotic resistance, virulence determinants, and metabolic prowess.
Flagella and Pili:
Numerous Eubacteria exhibit motility attributed to specialized extensions known as flagella, protruding from the cellular surface and facilitating directional movement towards or away from specific stimuli. Comprising the protein flagellin, flagella operate akin to propellers, engendering propulsive force essential for bacterial locomotion. Besides flagella, certain bacterial strains feature shorter appendages termed pili or fimbriae, pivotal for surface adhesion, biofilm establishment, and the process of bacterial conjugation, facilitating genetic exchange between individual cells.
Mesosomes and Inclusions:
In the cytoplasmic domain, Eubacteria may harbor mesosomes, intricately-folded invaginations of the plasma membrane, serving diverse roles encompassing cellular division, DNA replication, and respiratory processes. Furthermore, bacterial entities might amass storage granules or inclusions, comprising glycogen, lipids, sulfur, or polyphosphate, functioning as reservoirs for energy and vital nutrients. These repositories enable bacteria to endure periods of nutrient deprivation or environmental challenges, thereby ensuring their survival and resilience.
Eubacteria Types and Examples: Exploring Taxonomic Diversity
Major Phyla of Eubacteria
The taxonomic landscape of Eubacteria boasts a wealth of diversity, showcased through myriad phyla, each delineated by unique morphological, physiological, and ecological attributes. Within this taxonomic tapestry, certain phyla emerge as notable entities, distinguished by their ecological prominence and ubiquitous presence across diverse habitats.
Proteobacteria: Versatile and Abundant
The phylum Proteobacteria encapsulates a diverse array of Gram-negative bacterial taxa renowned for their metabolic adaptability and ecological omnipresence. Within the Proteobacteria lineage, multiple classes, notably Alpha-, Beta-, Gamma-, Delta-, and Epsilonproteobacteria, showcase unique metabolic proficiencies and ecological niches. Exemplars such as Escherichia coli, revered as a molecular biology paradigm, and Salmonella enterica, notorious for its etiological role in foodborne diseases, underscore the significance of this phylum in both scientific research and public health domains.
Firmicutes: Diverse and Functionally Versatile
The phylum Firmicutes stands as a prominent cohort within the domain of Gram-positive bacteria, celebrated for its extensive metabolic repertoire and ecological versatility. Encompassing a vast array of taxa, including Bacillus, Clostridium, Lactobacillus, and Staphylococcus, each member showcases distinctive physiological attributes and ecological adaptations. Bacillus subtilis, revered for its contributions to biotechnology and its status as a genetic model organism, serves as a quintessential example illustrating the functional breadth inherent within the Firmicutes phylum.
Actinobacteria: Prolific Producers of Bioactive Compounds
The phylum Actinobacteria stands as a diverse cohort of Gram-positive bacteria distinguished by their filamentous growth morphology and prolific synthesis of bioactive molecules. Within this group, genera such as Streptomyces, Mycobacterium, and Corynebacterium are prominent, renowned for their capacity to produce antibiotics, enzymes, and secondary metabolites of therapeutic interest. Streptomyces coelicolor, serving as a paradigmatic organism for investigating antibiotic biosynthesis, epitomizes the biosynthetic prowess of Actinobacteria and underscores their significance in pharmaceutical and biotechnological arenas.
Cyanobacteria: Oxygenic Photosynthesizers
Cyanobacteria, colloquially known as blue-green algae, represent a phylum of Gram-negative bacteria endowed with the ability to conduct oxygenic photosynthesis. These organisms assume pivotal roles within aquatic ecosystems, functioning as primary producers and thereby contributing substantially to carbon fixation and oxygen generation. Noteworthy genera encompass Anabaena, Spirulina, and Nostoc, which engage in symbiotic relationships with various organisms including plants, corals, and lichens, thus underscoring their ecological importance across diverse habitats.
Understanding the Distinctions: Eubacteria vs. Archaebacteria
Within the domain of microbiology, the systematic categorization of organisms into discrete domains serves as a cornerstone for elucidating their evolutionary affinities and biochemical attributes. Among the prominent domains within the prokaryotic realm, Eubacteria and Archaebacteria (Archaea) stand distinguished, each presenting distinctive traits and ecological habitats.
| Characteristic |
Eubacteria |
Archaebacteria |
| Cell Wall Composition |
Peptidoglycan/murein (NAG and NAM) |
Pseudomurein (NAT and NAG) |
| Cell Membrane Structure |
Phospholipids in a bilayer, unbranched |
Phospholipids in a monolayer, branched |
| Energy Source |
Obtains energy from Krebs cycle or glycolysis |
Unable to perform glycolysis or the Krebs cycle |
| Complexity |
Complex organisms |
Simple organisms |
| RNA Polymerase |
Simple |
Complex |
| Presence of Introns |
Absent |
Present |
| Membrane Lipid Configuration |
L-glycerol phosphate |
D-glycerol phosphate |
| Example Genera |
Bacillus, Mycobacterium, Clostridium |
Pyrobaculum, Ferroplasma, Lokiarchaeum, Thermoproteus |
Evolutionary Lineage and Phylogenetic Relationships
Eubacteria represents an extensive and diverse assemblage of microorganisms pervading nearly all ecosystems on our planet. They exhibit a wide spectrum of morphologies, metabolic pathways, and ecological functions, ranging from symbiotic mutualists to potent pathogens. A defining feature of Eubacteria is the presence of peptidoglycan within their cell walls and distinctive ribosomal RNA sequences, setting them apart from other domains of life.
In contrast, Archaebacteria, or Archaea, inhabit environments once deemed inhospitable to life, including extreme temperatures, pH levels, and salinity. Initially misclassified as bacteria owing to their prokaryotic cellular organization, Archaea showcase unique molecular and biochemical attributes, such as distinctive membrane lipids, DNA replication machinery, and histone-like proteins. Archaea are categorized into various phyla, each adapted to specific extreme habitats, with examples including halophiles thriving in high-salt conditions and thermophiles thriving in elevated temperatures.
Cell Wall Composition
A salient contrast between Eubacteria and Archaebacteria lies in the constitution of their cell walls. Eubacteria conventionally exhibit cell walls comprising peptidoglycan, a polymer characterized by repeating disaccharide units cross-linked by abbreviated peptides. This structural framework imparts rigidity and morphology to the cell, concurrently acting as a shield against osmotic stressors. In contrast, Archaebacteria diverge from this pattern by lacking peptidoglycan in their cell walls. Instead, they showcase a diverse array of cell wall compositions, exemplified by pseudomurein in select methanogens and proteinaceous S-layers in others.
Membrane Lipids
An additional notable disparity arises in the composition of membrane lipids. Eubacteria typically employ ester-linked phospholipids in their cell membranes, akin to those observed in eukaryotic organisms. These phospholipids are comprised of fatty acid chains tethered to a glycerol scaffold, forming a bilayer configuration. Conversely, Archaebacteria harbor distinct membrane lipids referred to as isoprenoid ethers or diethers, distinguished by branched hydrocarbon chains linked to glycerol through ether bonds. These unique lipid constituents contribute significantly to the robustness and resilience of Archaeal cell membranes, particularly in harsh environmental settings.
Metabolic Diversity
Both Eubacteria and Archaebacteria demonstrate notable metabolic versatility, embracing a diverse array of nutritional tactics and energy-yielding pathways. Eubacteria engage in a myriad of metabolic activities encompassing aerobic and anaerobic respiration, fermentation, and photosynthesis, endowing them with adaptability and resilience across diverse environmental milieus. Similarly, Archaebacteria exhibit metabolic plasticity, employing distinctive pathways such as methanogenesis, wherein methane serves as a metabolic byproduct in anaerobic habitats. Furthermore, certain Archaea demonstrate proficiency in chemosynthesis, harnessing energy derived from inorganic compounds to power cellular processes.
Adaptations to Extreme Environments
Archaebacteria, in particular, have developed specialized metabolic adaptations to flourish in extreme environments. Notably, Thermophilic Archaea thrive in environments characterized by high temperatures, such as hydrothermal vents and hot springs, where conventional enzymes would undergo denaturation. These organisms utilize thermophilic enzymes endowed with heightened stability and activity at elevated temperatures, facilitating the maintenance of metabolic functions under extreme thermal conditions. Similarly, halophilic Archaea have evolved to inhabit hypersaline environments by employing mechanisms to manage osmotic pressure and regulate ion concentrations within cells.


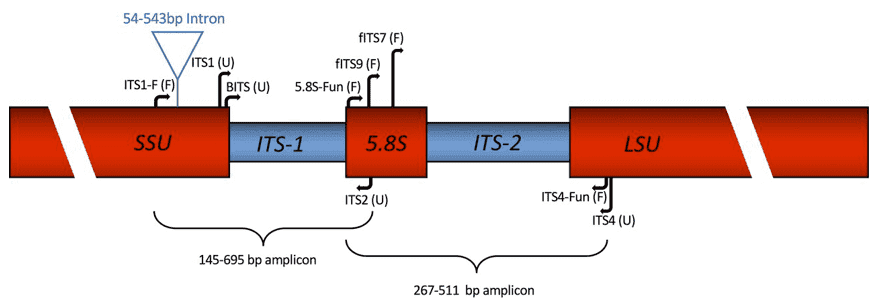
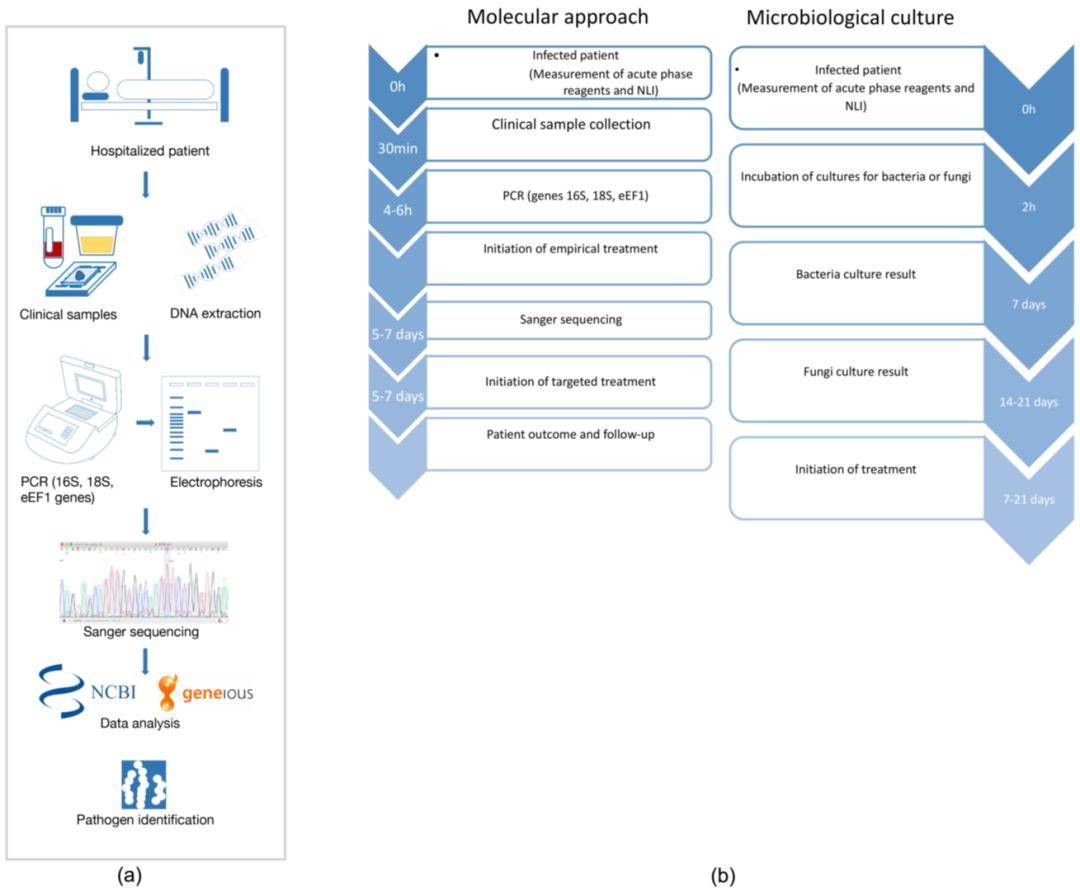
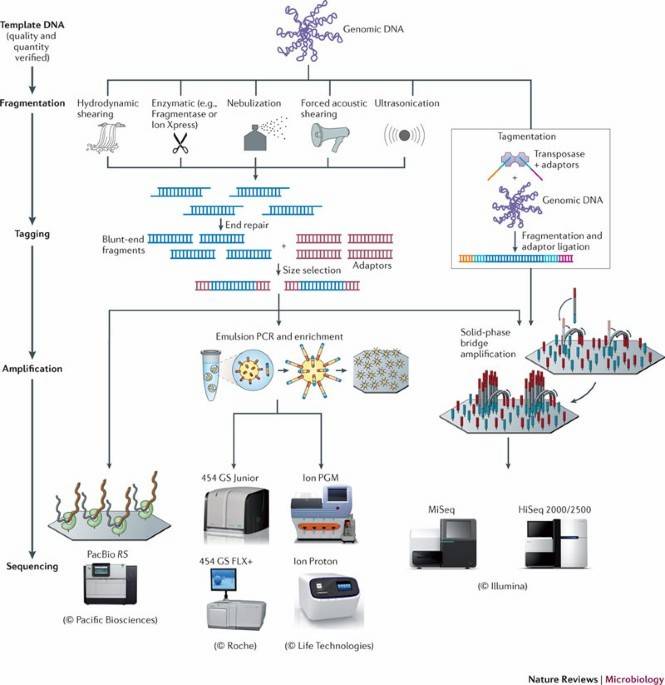 High-throughput bacterial genome sequencing
High-throughput bacterial genome sequencing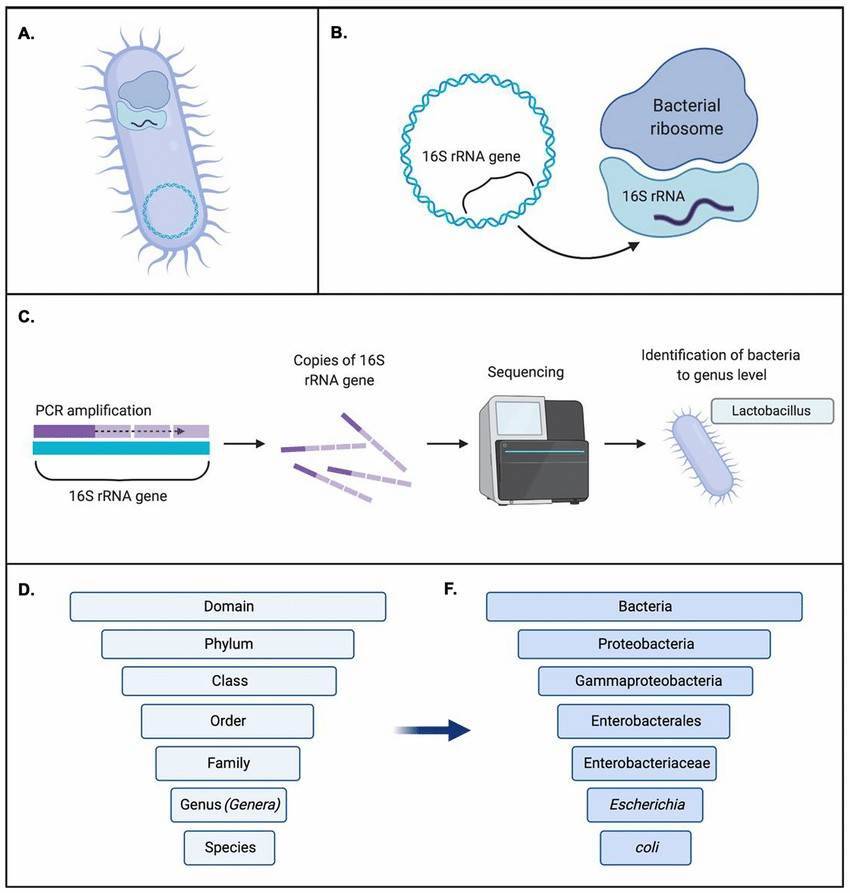 16S rRNA sequencing is the most common method used to analyze the bacteria(Sagar R. Patel et al, Current Urology Reports 2022)
16S rRNA sequencing is the most common method used to analyze the bacteria(Sagar R. Patel et al, Current Urology Reports 2022)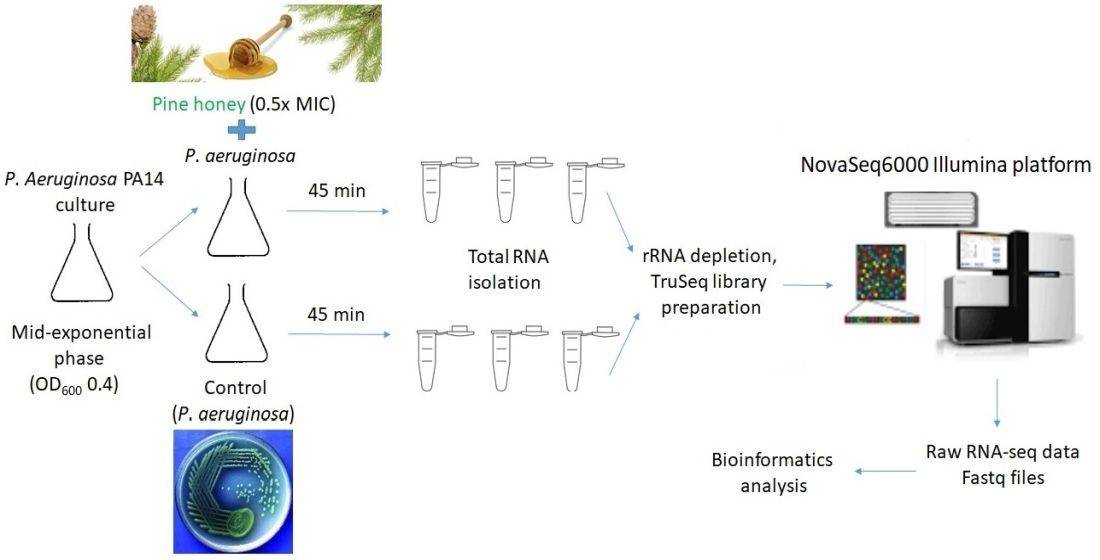 Transcriptomic Analysis of Pseudomonas aeruginosa Response
Transcriptomic Analysis of Pseudomonas aeruginosa Response