Exploring acRIP-seq: Decoding RNA Acetylation through Workflow, Analysis, and Future Perspectives
Similar to the widely studied epigenome, the "epitranscriptome" generated by post-transcriptional ribonucleoside modifications embeds a complex layer of regulation in RNA structure and function. Even though it has been shown that messenger RNA (mRNA) modifications can affect post-transcriptional metabolism by regulating mRNA stabilization, processing and/or translation, due to technical limitations, most of the studies to date have been based only on high-abundance transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs) rather than mRNAs. In addition, ac4C acetylation, one of the mRNA modifications, is even more difficult to detect because its modification abundance is at least 10-fold lower than that of m6A methylation, making it even more difficult to detect mRNA ac4C acetylation. The advent of acetylated RNA immunoprecipitation sequencing (acRIP-seq) as a protocol to identify acetylated mRNA regions on a transcriptome-wide scale has successfully broken this limitation.
acRIP-seq is a high-throughput sequencing tool for immunoprecipitation of intracellular RNA fragments with ac4C acetylation modifications by specifically recognizing ac4C-modifying antibodies. The emergence of low starting RNA amount acRIP-Seq technology has greatly reduced the cost of ac4C high-throughput assay, making it possible to detect ac4C distribution in a large number of clinical samples, and the initial ac4C assay was reported to require the use of 2 mg of total RNA, and after optimization of various technologies, it has been possible to achieve a starting amount of 150ug of total RNA.
With the improvement of the technology, ac4C-RNA acetylation modification will undoubtedly be another research hotspot after m6A RNA methylation modification, and the understanding of its workflow, analytical tools, application scope and future challenges will be the cornerstone of the ability to skillfully apply it to scientific research.
What is acRIP-seq and Why It Matters
acRIP-seq is a high-throughput sequencing (Next Generation Sequencing, NGS)-based technology for the comprehensive detection of the global map of N4-acetylcytosine (ac4C), an RNA modification. ac4C is a chemical modification conserved in eukaryotes and prokaryotes and plays an important role in numerous physiological processes. However, the chemical inertness of the acetylamino group in ac4C and its typical base pairing with guanosine make detection of the ac4C modification very difficult. Currently, commonly used ac4C sequencing methods include antibody immunoprecipitation-based sequencing (acRIP-seq) and sodium cyanoborohydride reduction-based sequencing (ac4C-seq).
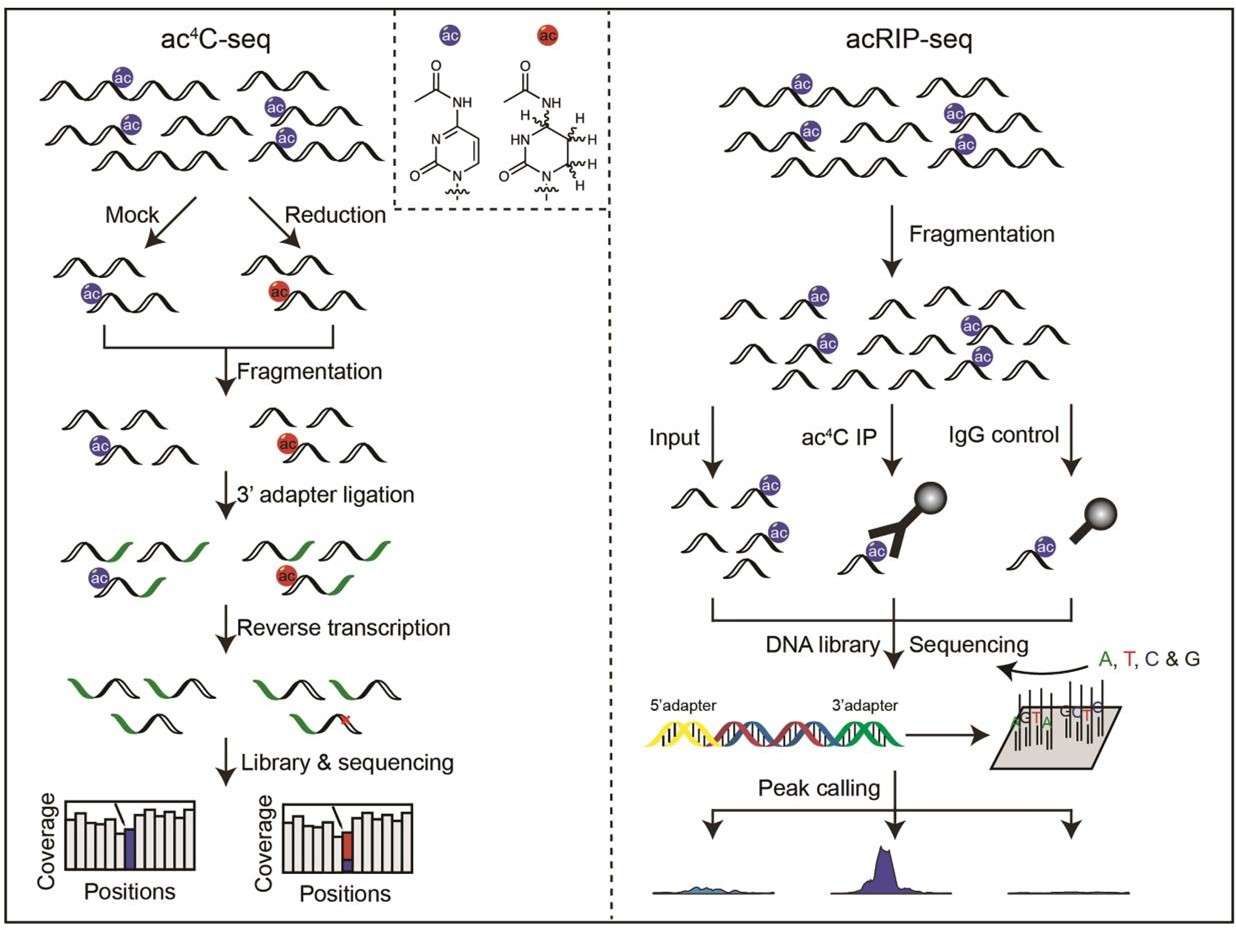 Scheme of ac4C-seq and acRIP-seq (Li et al., 2023)
Scheme of ac4C-seq and acRIP-seq (Li et al., 2023)
For ac4C-seq, RNA is first reduced or mock processed and then fragmented. After library construction and sequencing, ac4C is detected by analyzing C→T misincorporation sites. for acRIP-seq, RNA is first fragmented and then immunoprecipitated with an ac4C antibody or IgG.
In ac4C-seq, the high reactivity of sodium cyanoborohydride may lead to unwanted side reactions and, because of its complete dependence on the detection of C to T assays, may underestimate the abundance of RNA modifications during sequencing, thus failing to detect transcripts with lower abundance of ac4C modifications. In addition to this, the antibody-based acRIP-seq technique has the advantage of signal amplification compared to borohydride reduction-based methods, i.e., the use of ac antibodies to detect ac4C modified by 4C precipitation in RNA sequences and their deep sequencing, which has been used for the detection of ac4C in human and viral mRNAs.
Thus acRIP-seq has become the technique of choice for identifying ac4C modifications.
How acRIP-seq Works: Experimental Workflow
Based on the principle of antibody-specific binding to acetylated modified bases, acetylated RNA Immunoprecipitation (acRIP) is based on RNA immunoprecipitation to enrich for methylated modified fragments, followed by high-throughput sequencing to study acetylated RNA regions on a transcriptome-wide scale and obtain efficient results. Similar to meRIP-seq, acRIP-seq can be targeted to ac4C ornaments by simply switching to a commercially available antibody that recognizes ac4C.
Overview of the technical process
The acRIP-seq is a technology specifically designed to capture and analyze RNA molecules containing ac4C modified RNA. The process includes multiple steps including cell lysis and RNA extraction, RNA fragmentation, immunoprecipitation, RNA purification, library construction and high-throughput sequencing, which ultimately achieves precise localization and quantitative analysis of ac4C-modified RNA.
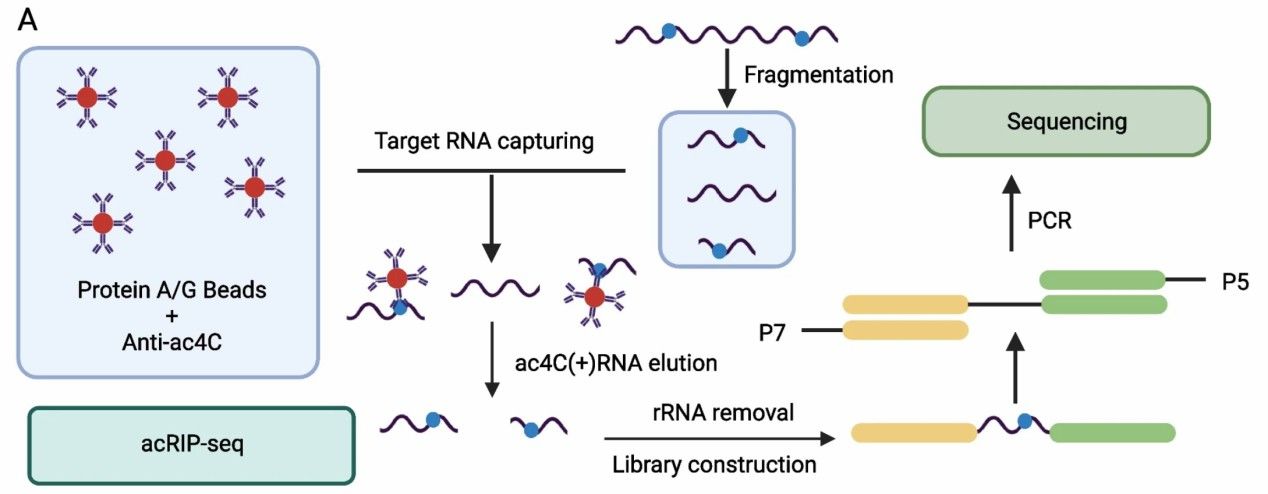 Workflow of acRIP-seq (Zhang et al., 2024)
Workflow of acRIP-seq (Zhang et al., 2024)
Cell Lysis and RNA Extraction
First, the cultured cell or tissue samples were lysed to form a single cell suspension. On the basis of lysis, the total RNA of the sample is extracted by RNA extraction kit. To minimize the interference of genomic DNA, the extracted RNA is digested by DNase enzyme to remove the residual DNA and ensure the purity of RNA.
RNA Quality Control and Quantification
Extracted RNA needs to be quality tested and quantitatively analyzed to ensure the validity of the experiment. The integrity of RNA is assessed using agarose gel electrophoresis or bioanalyzer to check for degradation. Meanwhile, RNA concentration is accurately measured by spectrophotometer or fluorescence quantification methods (e.g. Qubit) to provide data support for subsequent experiments.
RNA Fragmentation
To improve the efficiency of immunoprecipitation and subsequent sequencing, RNA needs to be broken into short fragments of approximately 100 nucleotides. RNA samples are usually fragmented by sonication or enzymatically. Subsequently, the size distribution of the RNA fragments is confirmed by agarose gel electrophoresis or capillary analysis to match the experimental requirements.
RNA Immunoprecipitation (RIP)
RNA immunoprecipitation (RIP), the core step of acRIP-seq, captures RNA fragments containing ac4C modifications by specific antibodies. RNA samples are incubated with an antibody that specifically recognizes ac4C, allowing it to bind to form an antibody-RNA complex. Next, these complexes are captured using agarose microbeads or magnetic beads encapsulated with Protein A/G, and RNA fragments containing ac4C modifications are isolated by centrifugation or magnetic racking.
RNA Elution and Purification
After immunoprecipitation, nonspecifically bound background RNA is removed by multiple washing steps.Subsequently, the antibody is digested with Protein A/G microbeads using proteinase K to release the captured RNA molecules. Finally, the RNA is purified by methods such as phenol-chloroform extraction or RNA purification columns to obtain high-purity ac4C-modified RNA.
Library Construction
The purified RNA samples need to be constructed into libraries suitable for high-throughput sequencing. First, ribosomal RNA is removed by rRNA removal reagents to further enrich for non-rRNA molecules. Next, the RNA fragments are ligated to sequencing junctions (e.g., P5 and P7) and reverse transcription is performed to synthesize cDNA. finally, the library is amplified by PCR to ensure the required sample volume for sequencing.
Sequencing
The amplified library can be used for high-throughput sequencing to obtain sequence information of ac4C-modified RNA. Commonly used platforms include high-throughput sequencing technologies such as Illumina, which enable high coverage and high precision analysis of RNA molecules.
Data Analysis: From Sequencing to Biological Insights
Data analysis, as the last step of acRIP-seq, provides us with a perspective from sequencing to biological knowledge, enabling us to identify ac4C-modified RNA regions by bioinformatics means, which mainly includes upstream and downstream analyses, such as quality control, comparison, and peak calling.
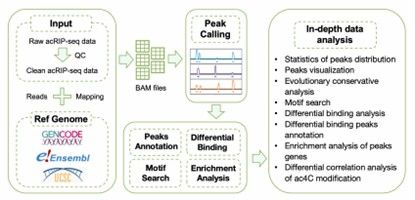 Bioinformatics Analysis Pipeline of acRIP-seq data.
Bioinformatics Analysis Pipeline of acRIP-seq data.
Data Preprocessing
In acRIP-seq experiments, raw sequencing data (usually in FASTQ format) are first acquired. Quality control (QC) of the data is performed by tools such as FastQC or Trim Galore to assess the quality of the sequencing data and remove low-quality bases (e.g., below Q30) as well as splice sequences. After QC is completed, high-quality clean data is generated to lay the foundation for subsequent analysis.
Read Mapping
Use tools such as STAR or Bowtie2 to compare clean data to a reference genome to ensure that reads are accurately localized to the corresponding region of the genome. Since acRIP-seq is usually strand-specific, strand information needs to be preserved to accurately localize the location of ac4C modifications. At the same time, the quality of the comparison results is assessed, including the comparison rate and the proportion of uniquely compared reads, to ensure that the data meets the analysis requirements.
Peak Calling and Visualization
Peak identification tools such as MACS2 or RIPSeeker were utilized to detect enriched peak regions, which correspond to ac4C modification sites on the RNA. Subsequently, genome browser tracks were generated using tools such as IGV or deepTools to visualize the enrichment of peaks. In addition, the distribution of peaks on gene features, such as promoters, exons, introns, and regions of the 3'UTR, was further analyzed to understand the distribution pattern of ac4C modifications.
Motif Search and Differential Binding Analysis
On the basis of peak identification, the consensus RNA sequence patterns in the enriched peaks were searched using tools such as HOMER or MEME to identify RNA sequence features associated with ac4C modification. Meanwhile, the difference in ac4C peak intensity between different experimental groups (e.g., control and treatment groups) was compared with the help of difference analysis tools such as diffBind or DESeq2. In addition, the differential peaks were annotated by tools such as ChIPseeker or bedtools to clarify their correspondence with gene features.
Functional and Enrichment Analysis
For the identified ac4C modification-related genes, gene ontology (GO) analysis was performed using tools such as clusterProfiler to reveal the biological processes, cellular components and molecular functions in which these genes are involved. Then, the signaling pathways affected by ac4C modification are identified by pathway enrichment analysis (e.g., KEGG, Reactome) to further explore their significance in biological functions. In addition, correlations between ac4C modifications and gene expression changes or other modifications can be analyzed to reveal the functional relevance of ac4C modifications.
Evolutionary Conservation Analysis
Comparative cross-species analysis of the conservation of ac4C peaks identifies RNA acetylation modification sites of evolutionary significance and reveals the functional role of ac4C modifications in species evolution.
Applications of acRIP-seq in Disease Research
As a cutting-edge RNA epitranscriptomics technology, acRIP-seq is capable of high-resolution detection of transcriptome-wide RNA modifications, in particular N4-acetylcytidine (ac4C), by antibody-mediated RNA immunoprecipitation coupled with sequencing. The development of this technology has provided a powerful tool for understanding the role of RNA modifications in disease pathogenesis. In recent years, studies have shown that ac4C modifications play a critical role in a variety of diseases. For example, in triple-negative breast cancer, acRIP-seq identified several ac4C-modified genes that serve as potential targeted therapeutic markers, providing new therapeutic targets for this aggressive cancer subtype. In addition, colorectal cancer studies have identified NAT10 as a key acetyltransferase that drives colorectal cancer progression by mediating ac4C modification to regulate the Wnt/β-catenin signaling pathway.
Further, acRIP-seq has shown important applications in the study of RNA modifications and chemotherapy resistance. In melanoma, NAT10-mediated ac4C modification acted on DDX41 mRNA to affect its expression level, which in turn caused chemoresistance and revealed possible drug targets. In neurodegenerative diseases, ac4C modification was found to enhance the translation efficiency of Vegfa mRNA, revealing a key molecular mechanism in the development of neuropathic pain. In addition, in cardiovascular and metabolic disease research, acRIP-seq revealed the impact of mRNA acetylation patterns on the vascular remodeling process and metabolic diseases, showing promise for the development of new therapeutic targets by targeting RNA modification pathways. These findings highlight the potential application of acRIP-seq technology in disease diagnosis, prognostic marker screening, and development of targeted RNA modification therapeutics, making it an indispensable tool in current biomedical research.
If you want to learn more about the acRIP-seq, please refer to:
Future Perspectives and Emerging Opportunities
The development of acRIP-seq technology has provided a powerful tool for resolving the role of RNA modifications in a wide range of diseases. nat10 is the first key enzyme found to catalyze RNA ac4C modifications, which are involved in a variety of pathological processes including cancer, neurodegenerative diseases, and cardiovascular diseases. NAT10 affects several key genes such as FSP1, PI3K-AKT pathway, and LINC00623 lncRNA by regulating ac4C modification, which promotes tumor proliferation, metastasis, and drug resistance. In addition, NAT10-mediated ac4C modification has been found to be associated with fatty acid metabolism, neuronal dysfunction, and metabolic diseases, demonstrating its broad biological significance.
Meanwhile, Remodelin, as a new molecule with anticancer activity, and the interaction of NAT10-mediated RNA modification with metabolic pathways provide new targets for disease diagnosis and treatment.The acRIP-seq technology is expected to further reveal these mechanisms and explore the application of RNA modification in disease diagnosis, prognosis prediction and personalized treatment, providing new ideas for cross-disciplinary disease research.
References:
- Arango, Daniel et al. "Immunoprecipitation and Sequencing of Acetylated RNA." Bio-protocol 9 12 (2019): e3278 . doi:10.21769/BioProtoc.3278
- Arango, Daniel et al. "Acetylation of Cytidine in mRNA Promotes Translation Efficiency." Cell175 (2018): 1872-1886.e24. doi:10.1016/j.cell.2018.10.030
- Li, Bin et al. "Transcriptome-wide profiling of RNA N4-cytidine acetylation in Arabidopsis thaliana and Oryza sativa." Molecular plant 16,6 (2023): 1082-1098. doi:10.1016/j.molp.2023.04.009
- Zhang, Shujun et al. "Recent advances in the potential role of RNA N4-acetylcytidine in cancer progression." Cell Communication and Signaling : CCS 22 (2024): 49. doi:10.1186/s12964-023-01417-5
! For research purposes only, not intended for clinical
diagnosis, treatment, or individual health assessments.
 Scheme of ac4C-seq and acRIP-seq (Li et al., 2023)
Scheme of ac4C-seq and acRIP-seq (Li et al., 2023) Workflow of acRIP-seq (Zhang et al., 2024)
Workflow of acRIP-seq (Zhang et al., 2024) Bioinformatics Analysis Pipeline of acRIP-seq data.
Bioinformatics Analysis Pipeline of acRIP-seq data.





