- Home
- Services
- Genome-wide DNA Methylation Analysis Service
- Super Enhancer Identification
- 5fC Sequencing
- Human & Mouse CpG Island Panel Sequencing
- Whole Genome Bisulfite Sequencing(WGBS)
- Reduced Representation Bisulfite Sequencing (RRBS)
- MeDIP Sequencing
- DNA 6mA Sequencing
- MRE-Seq Service
- Human DNA Methylation Microarray Service
- Long Read Sequencing DNA Methylation Service
- Mouse DNA Methylation Microarray Service
- MeDIP-qPCR & hMeDIP-qPCR
- RNA Modification Service
- TRAC-Seq
- RIP-qPCR
- ssDRIP-Seq
- DRIPc-seq
- ChIRP-Seq & ChIRP-MS
- PIRCh-seq
- SELECT-m6A Sequencing
- mRNA m5C BS-seq
- eCLIP-seq
- BID-Seq
- tRNA Modification Analysis
- miCLIP-seq
- GLORI-seq
- MeRIP-Seq
- m6Am Analysis Service
- m5c RNA-seq
- ONT Direct RNA Sequencing
- acRIP-seq & ac4C-seq
- 2'-O-methylated-seq Service
- O8G-Seq
- m1A RNA Methylation Analysis
- RNA m7G Methylation Sequencing
- PA-Ψ-seq
- GlycoRNA-seq Services
- MazF-qPCR m6A Detection
- MeRIP-qPCR
- DNA Hydroxymethylation Analysis Service
- Chromatin Analysis
- DAP-Seq
- Hi-C Service
- CUT&Tag Service
- ChIP-qPCR
- Genome-wide DNA Methylation Analysis Service
- Targeted DNA methylation analysis Service
- cfDNA Methylation Analysis
- Epigenomic Data Analysis
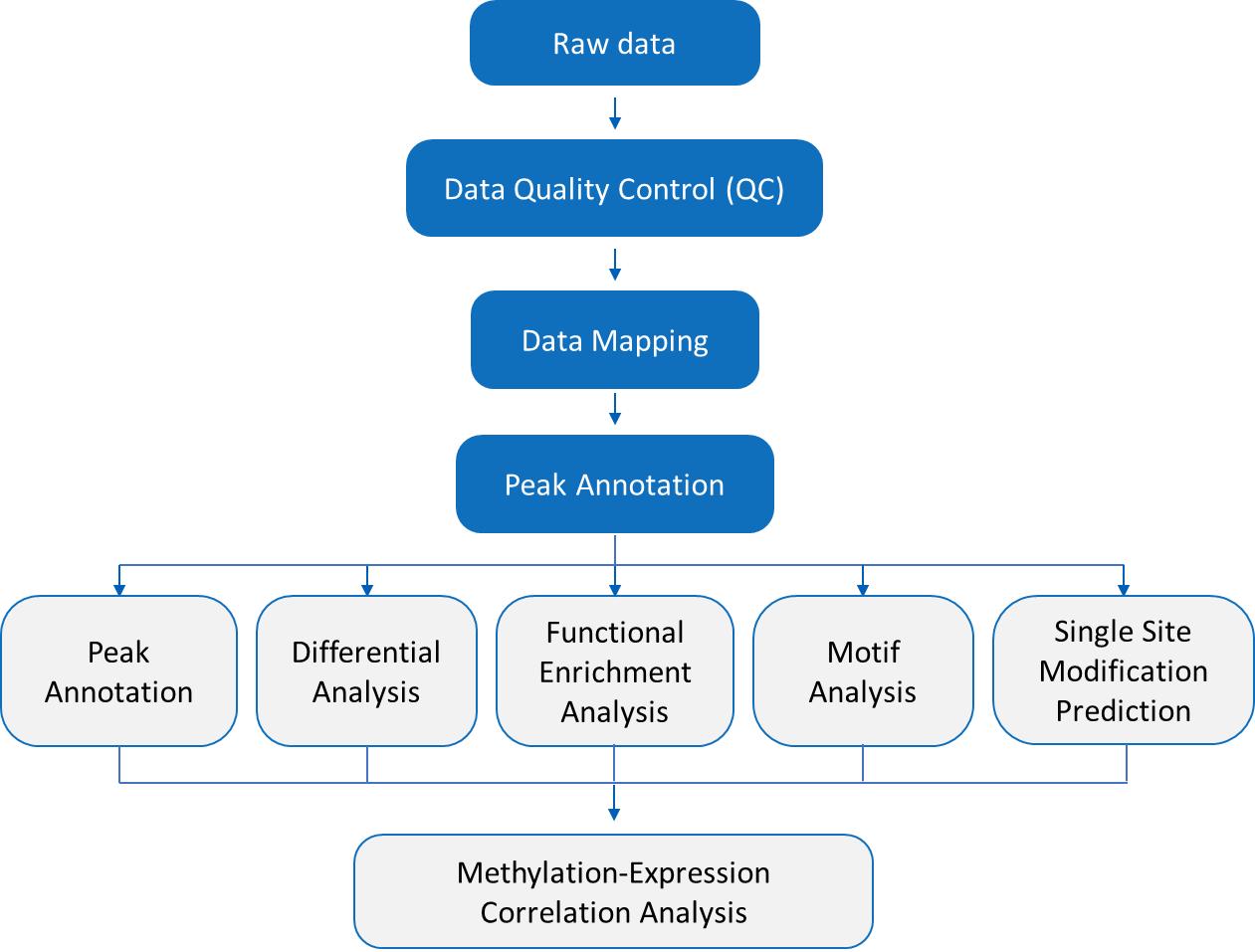
 m6Am-Exo-seq Dual IP Library Construction Workflow
m6Am-Exo-seq Dual IP Library Construction Workflow