We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy

RNA methylation represents a significant aspect of epigenetic research. Over 100 distinct RNA modifications have been identified, with methylation, hydroxymethylation, and acetylation among the most common. Among these, N6-methyladenosine (m6A) is the most prevalent methylation modification in RNA, with an average of 1 to 3 m6A sites per transcript. m6A modifications are found in the mRNA of most eukaryotes, including mammals, insects, plants, and yeast, as well as in certain viral RNAs. Additionally, m6A is present in transfer RNA (tRNA), ribosomal RNA (rRNA), small nuclear RNA (snRNA), and several long non-coding RNAs (lncRNAs).
RNA methylation is believed to play a crucial role in the regulation of gene expression, RNA splicing, RNA editing, RNA stability, and the control of mRNA lifespan and degradation.
m6A methylation refers to the selective addition of a methyl group to the nitrogen atom at the sixth position of adenosine residues within RNA molecules. This modification is catalyzed by the RNA methyltransferase complex (MTC), which comprises several core components, including METTL3, METTL14, and WTAP. The m6A-modified nucleotides are dynamic and reversible, undergoing demethylation by the action of the fat mass and obesity-associated protein (FTO) and AlkB homolog 5 (ALKBH5) enzymes.
The process of RNA methylation modification involves three primary classes of molecules: Writers, Erasers, and Readers.
Writers are methyltransferases that catalyze the addition of methyl groups to specific nucleotides within RNA, facilitating the modification process. The RNA methyltransferase complex, which includes METTL3, METTL14, and other associated proteins, is responsible for m6A methylation.
Erasers are demethylases that remove methyl groups from RNA, thus reversing the methylation process. Key enzymes in this category include fat mass and obesity-associated protein (FTO) and AlkB homolog 5 (ALKBH5), which specifically demethylate m6A-modified adenosines.
Readers are proteins that recognize and bind to methylated RNA, mediating the functional outcomes of the modification. These proteins, such as YTH domain-containing proteins, interpret methylation marks and influence RNA metabolism, including stability, splicing, and translation.
Together, these molecular participants regulate RNA methylation dynamics, thereby modulating gene expression and cellular function.
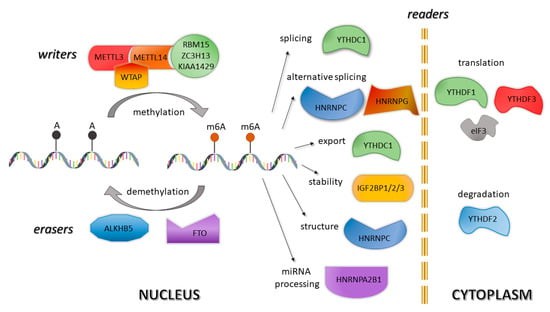 Figure 1. Roles of N6-methyladenosine "writers", "erasers" and "readers" in regulation of mRNA. (Anna Wardowska 2019)
Figure 1. Roles of N6-methyladenosine "writers", "erasers" and "readers" in regulation of mRNA. (Anna Wardowska 2019)
Due to its diverse physiological functions, m6A methylation is widely associated with numerous human diseases. Extensive research has revealed correlations between m6A and various cancers, including gastric cancer, prostate cancer, breast cancer, pancreatic cancer, renal cancer, mesothelioma, sarcoma, and leukemia. The depletion of METTL3, a core component of the m6A methyltransferase complex, has been shown to induce apoptosis in cancer cells and reduce their invasiveness. Hypoxia-induced activation of the demethylase ALKBH5 has been linked to the enrichment of cancer stem cells, further highlighting the role of m6A dynamics in oncogenesis.
Moreover, the fat mass and obesity-associated protein (FTO), a key regulator of energy metabolism, is closely associated with obesity. Studies have identified single nucleotide polymorphisms (SNPs) within the FTO gene that are significantly correlated with body mass index (BMI), obesity, and the development of type 2 diabetes mellitus. The involvement of m6A in neuronal disorders is also evident. For instance, neurodegenerative diseases may be influenced by m6A modifications, as FTO-regulated m6A methylation of critical transcripts has been implicated in homologous dopamine signaling pathways.
Additionally, knockdown or knockout of insulin-like growth factor 2 mRNA-binding proteins 1-3 (IGF2BP1-3) has been reported to decrease MYC protein expression, cellular proliferation, and colony formation in human cancer cell lines. ZC3H13, another member of the m6A methyltransferase complex, has been found to significantly inhibit colorectal cancer cell growth upon knockdown, underscoring the pivotal role of m6A in tumorigenesis.
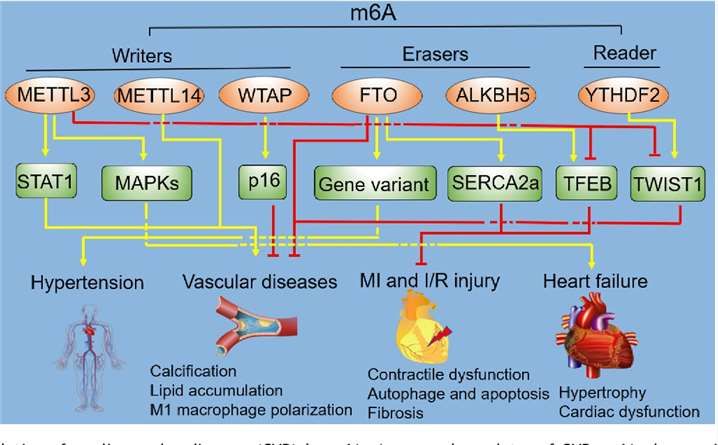 Figure 2. Regulation of cardiovascular diseases (CVD) by m6A. (Beijian Zhang et al,.2020)
Figure 2. Regulation of cardiovascular diseases (CVD) by m6A. (Beijian Zhang et al,.2020)
m6A methylation has been implicated in the regulation of viral infections, influencing the replication and infectivity of various RNA viruses. Notably, m6A methylation sites have been identified within the viral RNA genomes of several pathogenic viruses, including hepatitis viruses, adenoviruses, herpesviruses, Rous sarcoma virus, and influenza viruses. Recent studies have demonstrated that m6A modifications critically modulate the infection and replication efficiency of RNA viruses such as human immunodeficiency virus (HIV), hepatitis C virus (HCV), and Zika virus (ZIKV). These findings underscore the essential role of m6A and its associated factors in the regulation of viral life cycles and host-virus interactions.
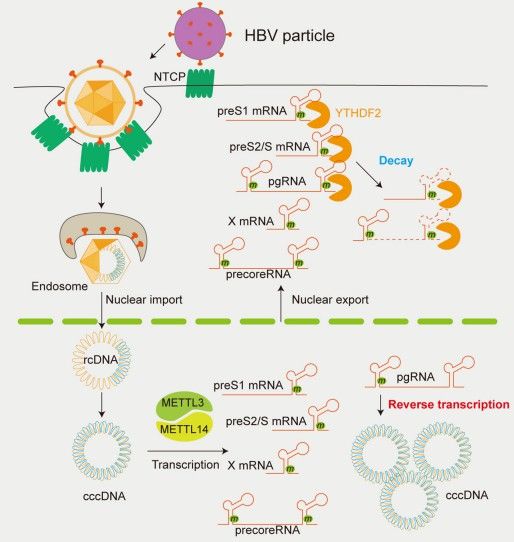 Figure 3. The role of m6A modification in differentially regulating the HBV life cycle. (Kim, GW. et al,. 2021)
Figure 3. The role of m6A modification in differentially regulating the HBV life cycle. (Kim, GW. et al,. 2021)
Numerous methodologies have been developed to detect m6A modifications, including methylated RNA immunoprecipitation sequencing (MeRIP-seq), m6A individual-nucleotide-resolution cross-linking and immunoprecipitation sequencing (miCLIP-seq), MeRIP-qPCR, and liquid chromatography-tandem mass spectrometry (LC-MS/MS).
MeRIP-seq utilizes immunoprecipitation (IP) to enrich m6A-modified RNA, coupled with sequencing analysis to generate m6A modification profiles. The approach includes two libraries: the IP-enriched library and the input library. The input library serves as an RNA-seq dataset, enabling gene expression analysis.
Technical Characteristics:
miCLIP-seq combines ultraviolet cross-linking with IP, enabling single-nucleotide resolution detection of m6A sites. By exploiting cytosine-to-thymine (CT) mutations, it accurately identifies m6A modification sites within a single peak.
Technical Characteristics:
MeRIP-qPCR is a targeted method for quantifying m6A modifications at specific regions, offering high accuracy. It necessitates the use of fully methylated and unmethylated standards.
Technical Characteristics:
Service you may intersted in
a. High-Throughput Sequencing for m6A Methylation Profiling
High-throughput sequencing techniques enable comprehensive mapping of m6A methylation patterns, including analyses of m6A peak counts, modified gene numbers, and peak distribution across gene elements. Additional analyses include motif identification and functional annotation of m6A-modified genes.
b. Screening of Differential m6A Peaks and Genes
Approaches include differential m6A peak identification, functional analysis of genes with differential m6A modifications, protein-protein interaction (PPI) analysis, and investigation of m6A modifications in candidate genes.
c. Integrative m6A Epitranscriptomics and Transcriptomics Analysis
Associative analysis between differential methylated genes (DMGs) and differential expressed genes (DEGs) can be performed, focusing on screening m6A-modified target genes.
a. Manipulation of m6A-Related Enzymes
Knockout or overexpression of m6A-associated enzymes combined with transcriptome sequencing facilitates the identification of target genes.
b. Functional Validation of Target Genes
Functional validation is achieved through knockout and overexpression studies of target genes.
c. Motif Mutagenesis for Direct Evidence
Motif-specific point mutations confirm direct regulation of target genes by m6A-related methyltransferases.
Terms & Conditions Privacy Policy Copyright © CD Genomics. All rights reserved.