We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy

RNA acetylation modification, as an emerging form of epigenetic regulation, exhibits important functions in regulating gene expression, translation efficiency and RNA stability. In particular, N4-acetylcytosine (ac4C) modification has become a hot spot of research in recent years due to its wide distribution and significant biological effects. With the development of acRIP-seq (acetylated RNA Immunoprecipitation sequencing) technology, researchers are able to accurately characterize the dynamics of RNA acetylation modifications and explore their mechanisms of action in a variety of diseases. This technology provides powerful support for understanding how RNA modifications drive disease onset, progression, and potential roles in therapy.
In cardiovascular diseases, acRIP-seq revealed how NAT10-mediated RNA modifications are involved in pathological processes by regulating cardiomyocyte iron death and cardiac remodeling. In cancer research, this technology has helped identify the close relationship between ac4C modifications and tumor proliferation, invasion, and prognosis, providing a new direction for precision treatment of tumors such as bladder and esophageal cancers. In addition, acRIP-seq has been applied in systemic lupus erythematosus (SLE), metabolic diseases, and neurodegenerative diseases, providing innovative perspectives for disease biomarker screening and therapeutic target research.
In this paper, we will discuss in detail the application of acRIP-seq technology in RNA acetylation modification research, covering representative cases in the fields of cardiovascular diseases, cancers and other systemic diseases, and then sort out the new discoveries of this technology in translational regulation, regulation of gene expression, and screening of disease markers, highlighting its significance in the promotion of precision medicine research.
Service you may intersted in
If you want to learn more about the acRIP-seq, please refer to:
As a state-of-the-art technology capable of detecting ac4C modification sites on a transcriptome-wide scale and revealing their roles in the regulation of gene expression, acRIP-seq has demonstrated a wide range of potential applications in cardiovascular disease, cancer, and other diseases, and offers new perspectives for understanding disease mechanisms and developing therapeutic strategies. In the following, we will discuss the application and importance of acRIP-seq in different diseases through specific case studies.
In the field of cardiovascular diseases, acRIP-seq has revealed the important role of RNA acetylation modifications in cardiac ischemia-reperfusion injury. For example, it was shown that NAT10-mediated modification of ac4C enhances the stability of Mybbp1a mRNA, which in turn activates the p53 signaling pathway and leads to cardiomyocyte iron death, thereby exacerbating cardiac injury. In addition, piRNA HAAPIR regulates cardiomyocyte apoptosis by promoting NAT10-mediated ac4C modification of Tfec mRNA, a finding that provides a new potential target for the treatment of myocardial infarction.
In cardiac ischemia/reperfusion (I/R) injury, N-acetyltransferase 10 (NAT10)-mediated modification of RNA acetylation has an important impact on cardiomyocyte iron death. It was shown that during I/R, p53 transcription activates NAT10, which enhances the stability of Mybbp1a mRNA through ac4C modification, which in turn activates the p53 signaling pathway and inhibits the transcription of the anti-iron deposition gene, SLC7A11, leading to increased cardiomyocyte iron death. Furthermore, it was found that inhibition of NAT10 expression and function significantly attenuated myocardial iron death and I/R injury. These findings provide new ideas and therapeutic targets for the clinical management of ischemic heart disease.
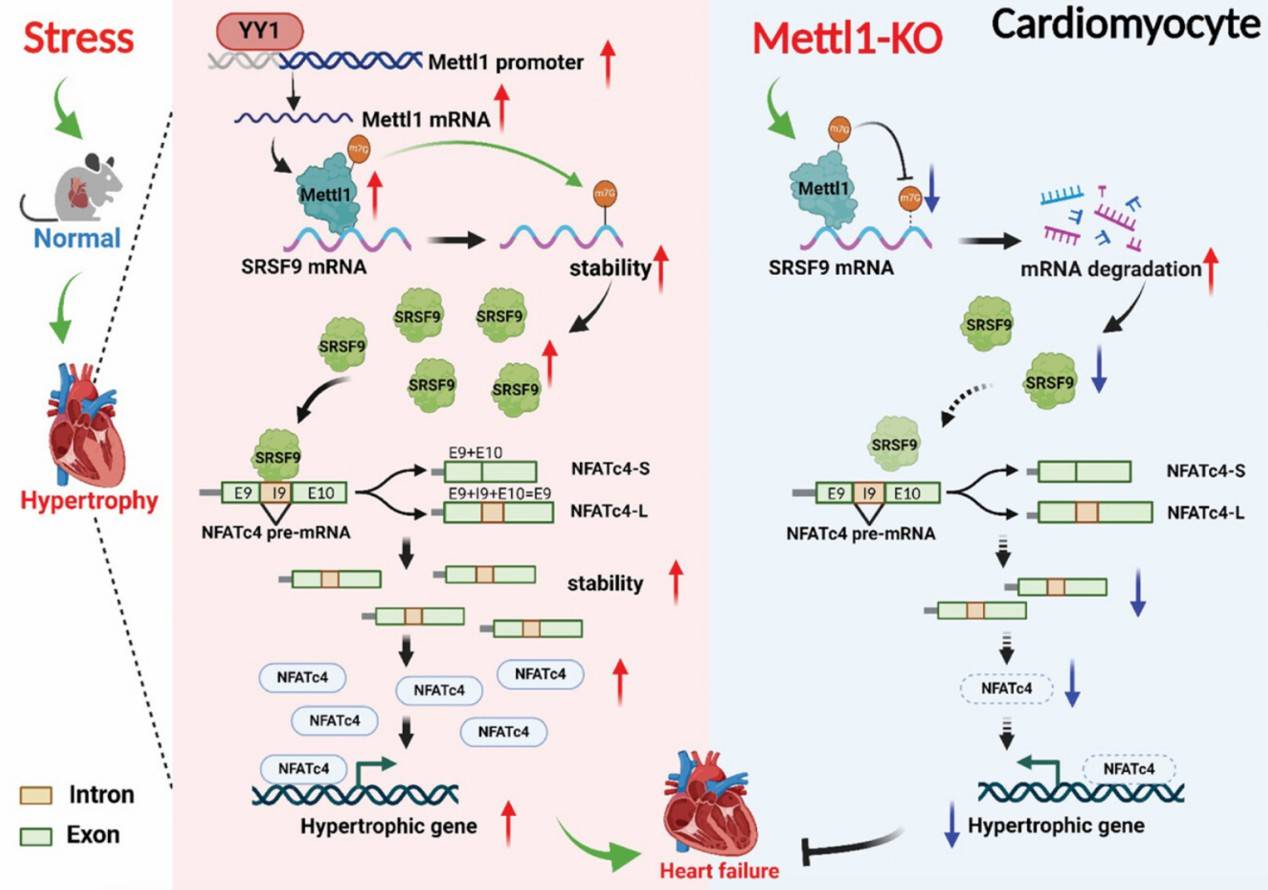 M7G methyltransferase targets the SRSF9/NFATc4 axis to regulate cardiac hypertrophy (Yu et al., 2024)
M7G methyltransferase targets the SRSF9/NFATc4 axis to regulate cardiac hypertrophy (Yu et al., 2024)
During cardiac remodeling, NAT10-mediated ac4C acetylation modification is involved in pathological processes such as cardiomyocyte hypertrophy and fibrosis by altering the expression of key genes. Using acRIP-seq technology, researchers revealed the ac4C acetylation modification profile in a cardiac remodeling model and found that NAT10-regulated genes such as Cd47 and Rock2 have important roles in cardiac remodeling. Specifically, NAT10 improves the stability and translational efficiency of Cd47 and Rock2 transcripts by upregulating their mRNA ac4C modifications, leading to their increased protein expression during cardiac remodeling. Inhibition of NAT10 expression or function significantly ameliorated cardiac dysfunction and fibrosis in cardiac remodeling models. In addition, the use of Remodelin, a NAT10 inhibitor, in a TAC model of cardiac remodeling further demonstrated that Remodelin significantly ameliorated cardiac dysfunction, cardiomyocyte hypertrophy, fibrosis, and inflammatory responses, whether administered before or after TAC induction. These studies provide potential new targets for the treatment of cardiac remodeling in heart failure.
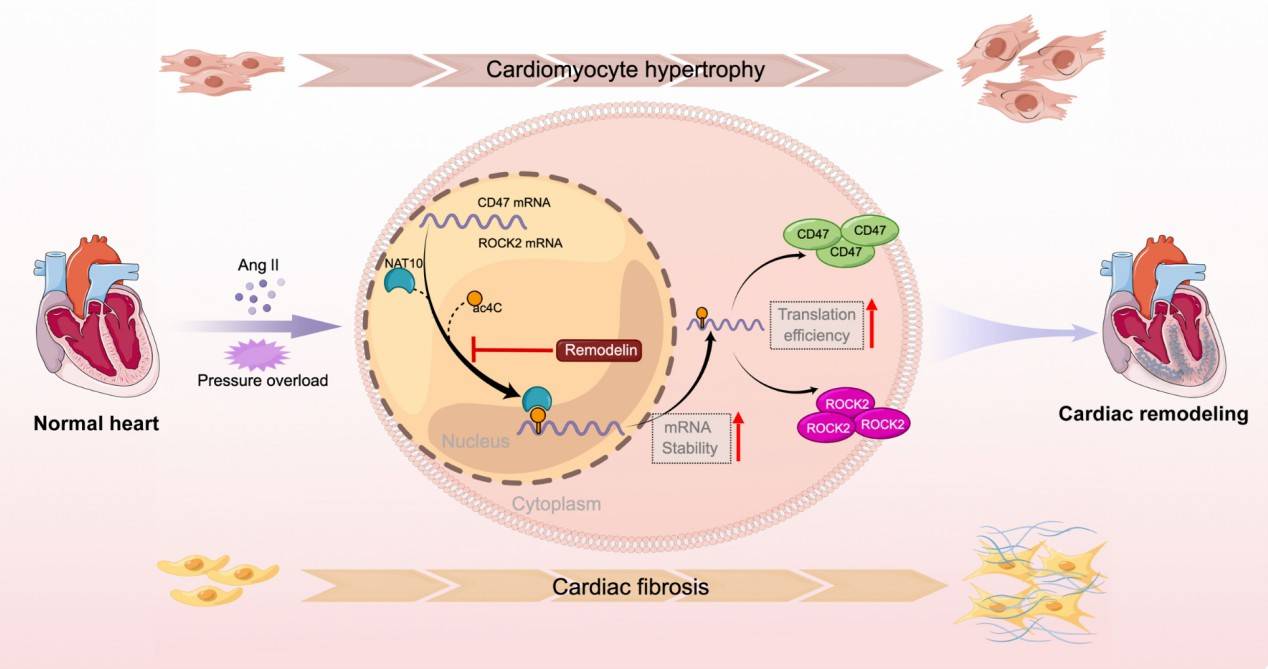 NAT10 is involved in cardiac remodeling through ac4C-Mediated transcriptomic tegulation (Shi et al., 2023)
NAT10 is involved in cardiac remodeling through ac4C-Mediated transcriptomic tegulation (Shi et al., 2023)
In cancer research, the application of acRIP-seq has helped reveal the regulatory mechanisms of RNA acetylation modifications in tumor cell proliferation and migration. By detecting changes in ac4C modifications in tumor cells, researchers were able to identify genes and signaling pathways associated with tumor progression, providing new ideas for precision cancer treatment.
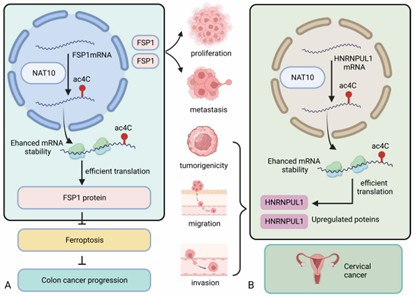 NAT10-related mechanisms involved in tumor progression (Zhang et al., 2024)
NAT10-related mechanisms involved in tumor progression (Zhang et al., 2024)
In gastric cancer studies, acRIP-seq revealed that mRNA ac4C modification leads to hyperactivation of the HIF-1 pathway through a NAT10/SEPT9/HIF-1a positive feedback loop, induces glycolytic addiction, and improves hypoxia tolerance in gastric cancer cells. This mechanism suggests a correlation between mRNA ac4C modification and anti-angiogenic treatment resistance. Based on this, the researchers proposed a strategy to overcome therapeutic resistance by combining an anti-angiogenic drug (e.g., apatinib) with an ac4C inhibitor (e.g., Remodelin). This combined treatment regimen is able to block glucose metabolic reprogramming while reducing tumor blood supply, enhancing the anti-tumor effect and providing a new idea for improving gastric cancer treatment.
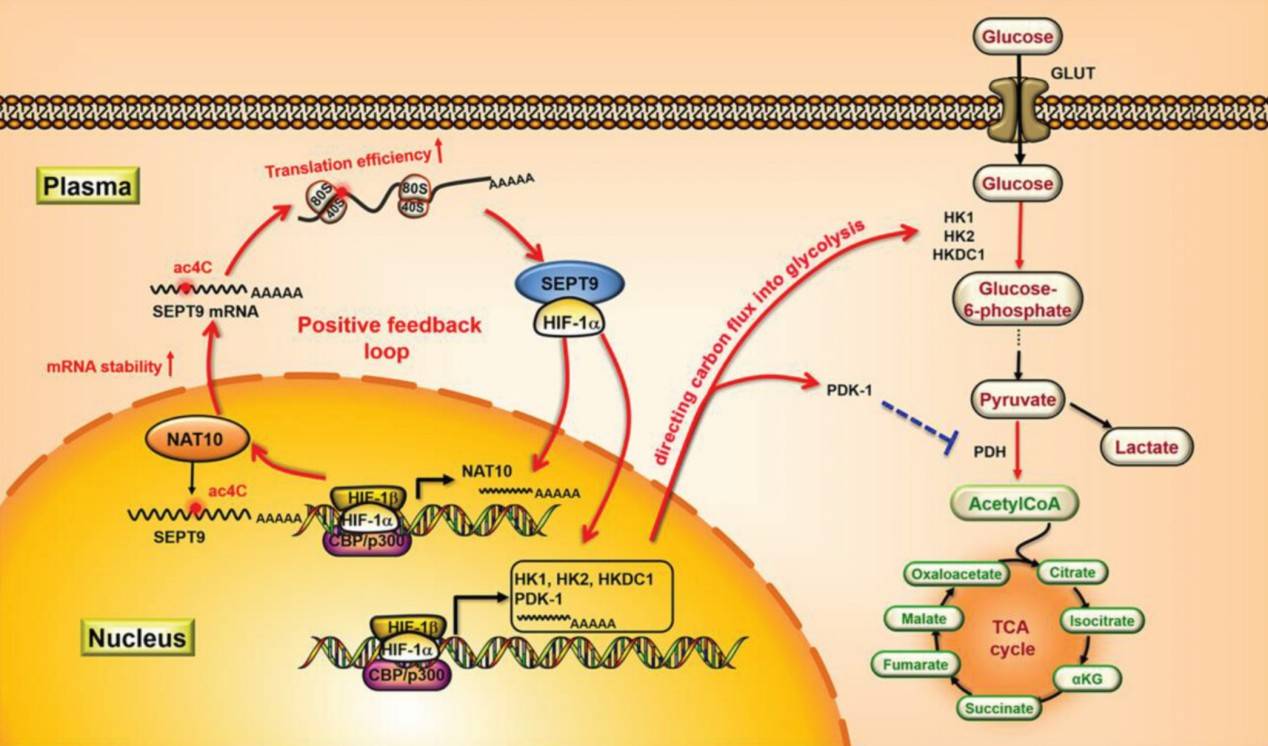 Schematic diagram depicting the mechanism of NAT10/SEPT9/HIF-1α positive feedback loop regulating glycolysis addiction in GC cells (Yang et al., 2023)
Schematic diagram depicting the mechanism of NAT10/SEPT9/HIF-1α positive feedback loop regulating glycolysis addiction in GC cells (Yang et al., 2023)
In bladder cancer studies, N-acetyltransferase 10 (NAT10) was found to promote tumor cell proliferation and migration by catalyzing ac4C modifications in target transcripts. Specifically, NAT10 directly binds to ac4C-rich targets such as BCL9L, SOX4 and AKT1. The expression of these genes is closely associated with tumor aggressiveness, migration ability and progression. For example, high expression of BCL9L was positively correlated with tumor aggressiveness, migratory ability, and tumor progression, whereas patients with its low expression had significantly longer survival times than those with its high expression. In addition, SOX4 expression was positively correlated with tumor invasiveness, and patients with high expression were more likely to develop lymph node metastasis.
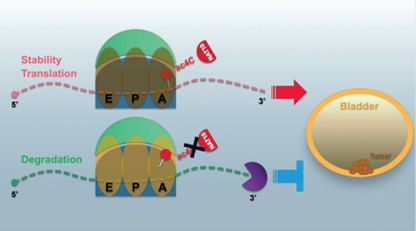 NAT10-mediated mRNA N4-acetylcytidine modification promotes bladder cancer progression (Wang et al., 2022)
NAT10-mediated mRNA N4-acetylcytidine modification promotes bladder cancer progression (Wang et al., 2022)
In addition, acRIP-seq has shown a wide range of applications in the study of other diseases. For example, in metabolic diseases, RNA acetylation modifications have been found to affect insulin signaling and adipocyte function, which in turn are involved in the pathological processes of diseases such as diabetes and obesity. In neurodegenerative diseases, RNA acetylation modifications may affect disease progression by regulating gene expression and protein translation efficiency in neurons.
In studies of SLE, acRIP-seq was used to map ac4C acetylation throughout the transcriptome in CD4+ T cells from SLE patients. It was found that the distribution of ac4C in mRNA transcripts was highly conserved and predominantly enriched in coding sequence regions of mRNAs. This finding reveals a potential mechanism for the role of RNA acetylation modifications in SLE and provides new insights into the understanding of the immunoregulatory abnormalities of the disease.
In viral infection studies, acRIP-seq has helped reveal the role of RNA acetylation modifications in viral replication and pathogenicity. For example, it was found that an ac4C modification catalyzed by the host acetyltransferase NAT10 is present in the 5'UTR region of the enterovirus 71 (EV71) genome. This modification enhances translation of viral RNA by selectively recruiting PCBP2 to IRES and improves RNA stability. The discovery of this mechanism provides a new target for the development of antiviral therapeutic strategies.
The application of acRIP-seq in the study of neurodegenerative diseases has revealed the important role of RNA acetylation modifications in neuronal function and disease progression. It was shown that RNA acetylation modifications may affect the pathological process of neurodegenerative diseases by regulating gene expression and protein translation efficiency in neurons.
These studies demonstrate the wide application of acRIP-seq in a variety of diseases, reveal the important role of RNA acetylation modification in disease occurrence and development, and provide new ideas and strategies for disease diagnosis and treatment.
In recent years, acRIP-seq has made significant progress in several areas of research. First, in translation regulation, RNA acetylation modification has been found to significantly affect the binding of mRNA to ribosomes, which in turn regulates the efficiency of protein synthesis. Second, in gene expression regulation, RNA acetylation modifications further regulate the diversity and complexity of gene expression by affecting mRNA stability and variable splicing. Finally, in the screening of disease biomarkers, acRIP-seq reveals specific changes of RNA acetylation modifications in a variety of diseases, providing new potential targets for early diagnosis and treatment of diseases.
RNA acetylation modifications play an important role in regulating protein translation efficiency. It has been shown that ac4C modification enhances the translation efficiency of mRNAs, a process that is mainly realized by increasing the binding capacity of mRNAs to ribosomes. For example, in studies of enterovirus 71 (EV71), ac4C modification enhanced the translation of viral RNA by selectively recruiting PCBP2 to IRES and improved RNA stability. In addition, the ac4C modification was able to influence the function of tRNAs, which in turn affects the protein synthesis process.
RNA acetylation modification plays an important role in the regulation of gene expression. NAT10, as a major ac4C modifying enzyme, regulates mRNA stability and translational efficiency through acetylation modification, thereby affecting gene expression. For example, in systemic lupus erythematosus (SLE), acRIP-seq revealed ac4C acetylation profiles throughout the transcriptome in CD4+ T cells from SLE patients and found that the distribution of ac4C in the mRNA transcripts was highly conserved and enriched with mRNA coding sequence regions. In addition, ac4C modifications were able to influence variable splicing of RNAs, further regulating the diversity and complexity of gene expression.
RNA acetylation modifications show great potential in the screening of disease biomarkers. With acRIP-seq technology, researchers are able to detect ac4C modification sites on a transcriptome-wide scale, thereby identifying specific RNA modifications associated with diseases. For example, in pancreatic cancer research, LINC00623 lncRNA binds to NAT10, promoting the formation of the ac4C modification in mRNAs, enhancing gene expression and leading to tumorigenesis. Detection of this modification provides new biomarkers for early diagnosis and treatment of the disease.
The emergence of acRIP-seq technology has revolutionized the study of RNA acetylation modification, offering an unprecedented ability to map and analyze this critical epitranscriptomic mark. By unveiling its essential roles in translational regulation, RNA stability, and gene expression control, acRIP-seq has significantly advanced our understanding of fundamental biological processes. This technology has also highlighted RNA acetylation as a key player in diverse cellular functions, laying the groundwork for further exploration of its broader biological significance.
In disease research, acRIP-seq has proven instrumental in uncovering the involvement of RNA acetylation in the pathogenesis of conditions such as cardiovascular diseases, cancer, and neurodegenerative disorders. Its ability to identify acetylation-related biomarkers has opened new avenues for early diagnosis, prognosis, and the development of targeted therapies. By shedding light on these mechanisms, acRIP-seq not only enhances the understanding of disease biology but also supports the advancement of precision medicine through innovative therapeutic strategies.
As research continues to deepen, RNA acetylation modification is poised to become a pivotal focus within the field of epitranscriptomics. With the ongoing optimization of acRIP-seq and its integration with other omics technologies, the potential to map acetylation across various biological contexts will further amplify its impact. This progression promises to bridge the gap between fundamental science and clinical application, unlocking new opportunities for improving human health and combating complex diseases.
References
Terms & Conditions Privacy Policy Copyright © CD Genomics. All rights reserved.