Whole genome analysis is a comprehensive concept that encompasses all research methods related to the generation, processing, and interpretation of data for the entire genome. This includes, but is not limited to, whole genome sequencing (WGS), genome-wide association studies (GWAS), and whole genome linkage analysis. This article provides a comprehensive introduction to several of these research methods and related sequencing technologies, with the aim of offering a thorough knowledge framework.
What is Genome-Wide Analysis
Genome-wide analysis represents a suite of methodologies designed to assess genetic variations and patterns across entire genomes. By systematically examining these variations, researchers gain profound insights into the genetic underpinnings of complex traits and diseases. This field has revolutionized our understanding of genetics and genomics, enabling the discovery of associations between genetic variants and phenotypic traits.
Service you may intersted in
Resource
Key Approaches in Genome-Wide Analysis
Genome-Wide Association Studies (GWAS)
GWAS focus on identifying single-nucleotide polymorphisms (SNPs) associated with specific diseases and traits. By comparing genetic data from large populations, GWAS has uncovered thousands of genetic variants linked to complex traits. This approach is invaluable for understanding the genetic basis of diseases like diabetes, cancer, and cardiovascular disorders.
GWAS analysis involves five main steps, from materials to results: sample collection, genotyping, phenotype investigation, association analysis, and functional validation.
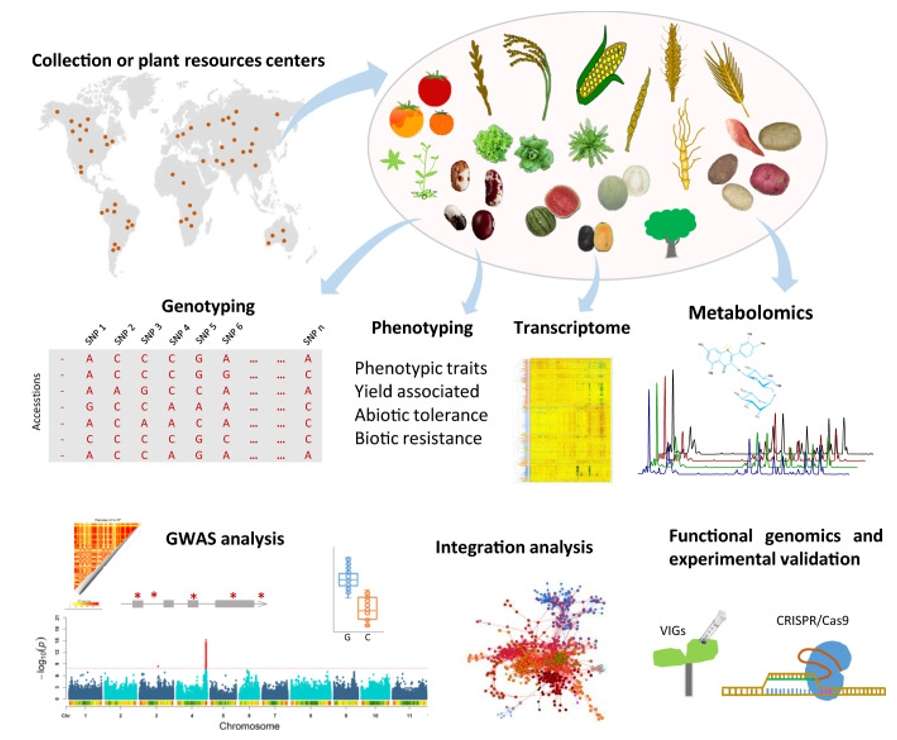 Fig. 1. A schematic view of GWAS in plants.(Alseekh, et.al, 2021).
Fig. 1. A schematic view of GWAS in plants.(Alseekh, et.al, 2021).
For those new to GWAS analysis, sample selection and phenotype data collection are especially crucial. Regarding sample selection, the recommended population size is typically at least 200 individuals, and there should be no obvious subpopulation differentiation among the samples. If subpopulations exist, it is recommended that each subpopulation contains more than 200 samples. As for phenotype data, accurate phenotypic measurements are key, and traits with higher heritability should be prioritized. Phenotype data can be classified into three categories: quantitative traits (controlled by multiple genes, normally distributed, and can be analyzed by growing or breeding under consistent environmental conditions or by synthesizing data from multiple years and locations), qualitative traits (controlled by a single gene, represented as binary values such as 0 or 1), and ordinal traits (controlled by multiple genes, phenotypically similar to qualitative traits, such as resistance traits, requiring precise measurement data for each individual).
Genome-Wide Linkage Analysis
Linkage analysis is a traditional yet essential tool used to locate genes associated with inherited traits. By analyzing the co-segregation of genetic markers with traits within families, this method is particularly effective for studying rare diseases. Linkage analysis is often used in conjunction with filtering methods in WGS, playing a crucial role in identifying rare variants, where GWAS may have limitations. Genome-wide linkage analysis continues to play a pivotal role in uncovering the genetic basis of Mendelian disorders.
A study investigated the genetic basis of attention deficit hyperactivity disorder (ADHD) in a Dutch family with eight affected individuals, five of whom had additional psychiatric disorders. Genome-wide linkage analysis identified two genomic regions, on chromosomes 7 and 14, showing significant allele sharing among affected family members. The regions 7p15.1–q31.33 and 14q11.2–q22.3 were confirmed to share the same haplotype across all patients(Vegt, et.al, 2021).
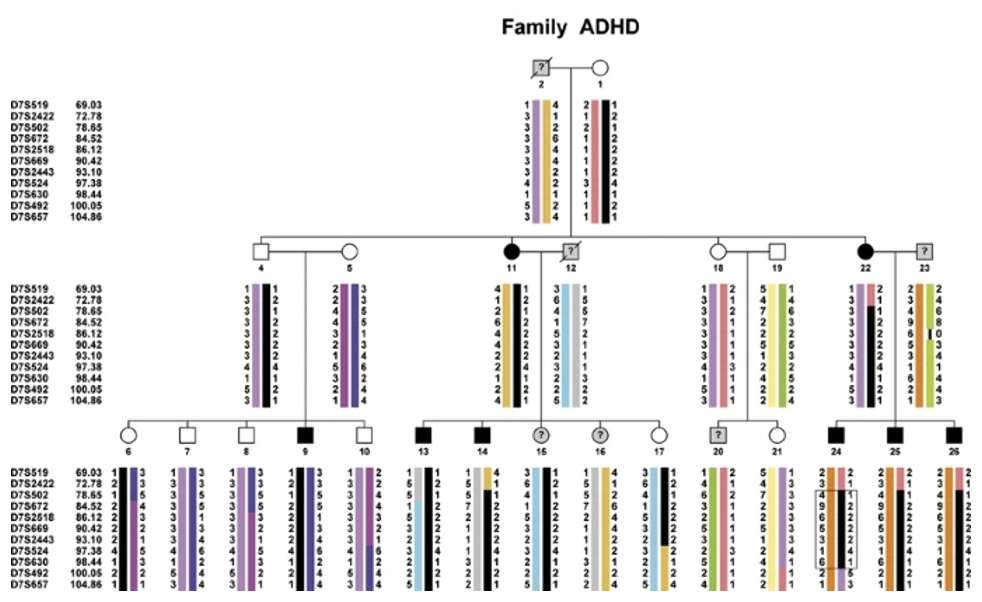 Fig. 2. Family tree showing microsatellite haplotypes.(Vegt, et.al, 2010)
Fig. 2. Family tree showing microsatellite haplotypes.(Vegt, et.al, 2010)
Genome-Wide Methylation Analysis
DNA methylation, a crucial epigenetic modification, regulates gene expression and influences various biological processes. Genome-wide methylation analysis examines these modifications, providing insights into gene regulation, oncology, developmental biology, and aging. Advanced methods like whole-genome bisulfite sequencing facilitate detailed mapping of methylation patterns across the genome, offering a deeper understanding of epigenetic regulation.
The core of DNA methylomics research lies in the mining of DNA methylation data, which generally involves three steps. First, a comprehensive analysis of genome-wide methylation changes is conducted, including the analysis of average methylation level changes, distribution changes, dimensionality reduction, clustering, and correlation analysis. Second, differential methylation analysis is performed to identify specific differentially methylated genes, including the identification of DMCs/DMRs/DMGs, the distribution of DMCs/DMRs across genomic elements, TF binding analysis, temporal methylation data analysis strategies, and functional analysis of DMGs. Lastly, DNA methylomics and transcriptomics are integrated for association analysis, including meta gene-wide associations, DMG-DEG correspondences, and network associations.
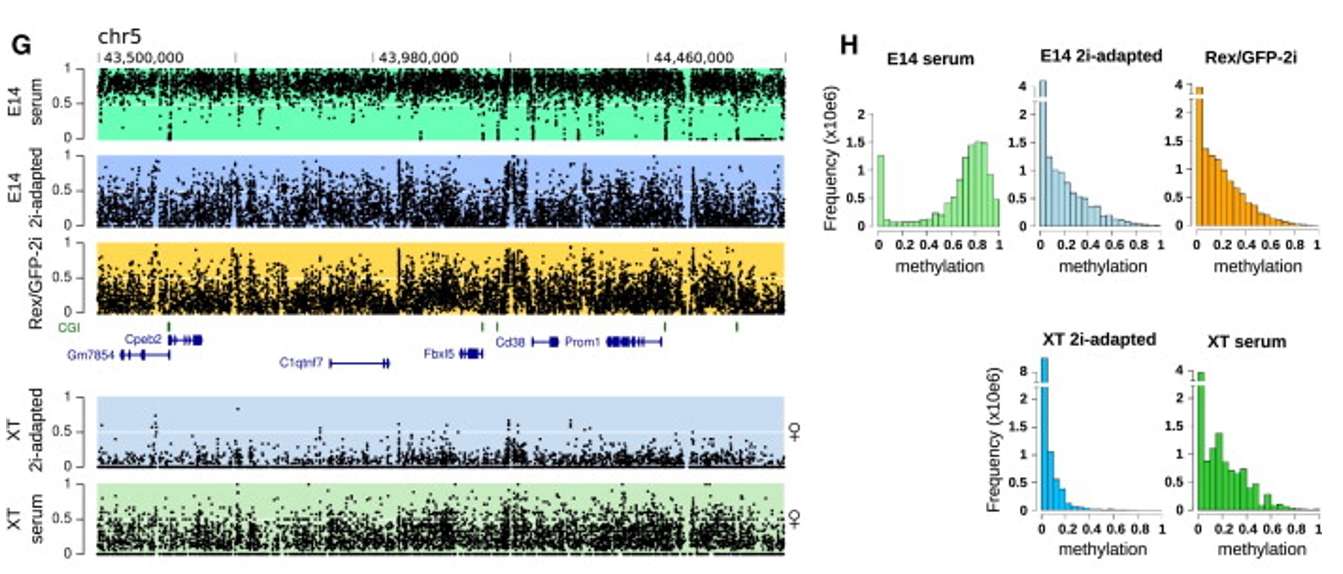 Fig. 3.Comparison of methylation modification of mouse embryonic stem cells in different states.(Habibi, et.al, 2013)
Fig. 3.Comparison of methylation modification of mouse embryonic stem cells in different states.(Habibi, et.al, 2013)
Service you may intersted in
Genome-Wide Transcriptome Analysis
Transcriptome analysis examines the entire set of RNA transcripts in a cell or tissue by RNA sequencing, shedding light on gene expression under different conditions. This approach is integral to understanding disease mechanisms, cellular responses, and plant biology. By identifying differential gene expression patterns, genome-wide transcriptome analysis has become a cornerstone for studying cancer biology, immune responses, and crop improvement.
A study selected two vegetable soybean varieties with different seed hardness to investigate the factors influencing seed hardness. Transcriptome analysis was performed at five stages of seed development. The analysis revealed that, during the middle and late stages of seed development, genes involved in the synthesis or degradation of starch, storage proteins, and fatty acids were differentially expressed between the two varieties, leading to differences in the accumulation of stored substances during seed maturation(Wang,et.al,2024).
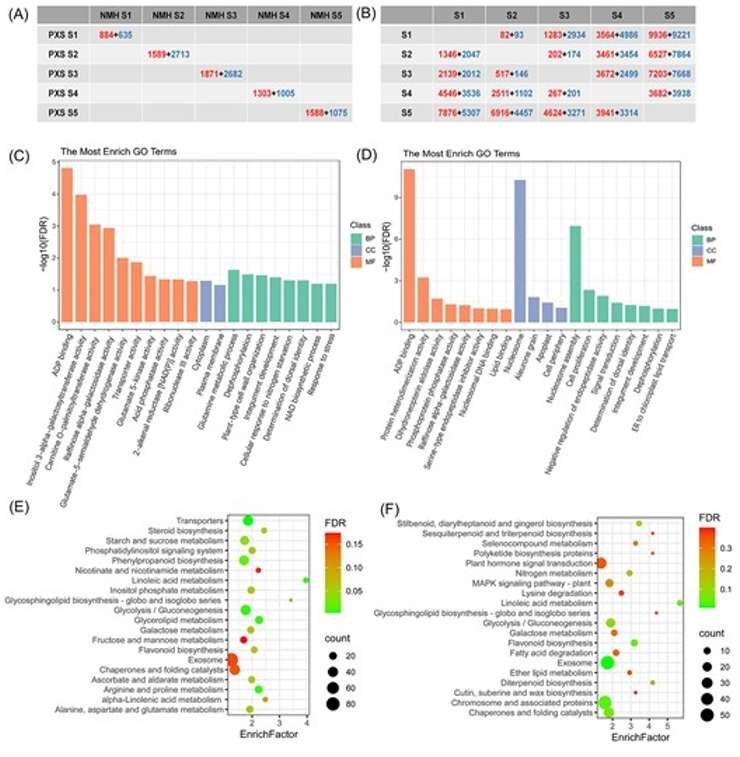 Fig. 4. Differentially expression genes in different samples.(Wang,et.al,2024)
Fig. 4. Differentially expression genes in different samples.(Wang,et.al,2024)
Service you may intersted in
Genome-Wide Analysis in Plants
Genome-wide analysis has transformative applications in plant biology, from understanding genetic diversity to improving crop resilience. By identifying genes responsible for desirable traits like drought tolerance and disease resistance, genome-wide studies contribute to sustainable agriculture. Combining GWAS, transcriptomics, and methylation studies, plant genomics continues to unlock pathways for crop optimization. See more in our genome wide analysis in plants article.
Methodologies and Technologies
High-Throughput Sequencing Techniques
Cutting-edge sequencing platforms, including Illumina, PacBio, and Oxford Nanopore, enable the efficient analysis of vast genomic datasets. These technologies underpin most genome-wide studies, offering unparalleled accuracy and throughput. For example, whole-genome sequencing provides comprehensive insights into genetic variations, supporting discoveries in both human and plant genomics.
Microarray Technologies
Microarrays remain a cost-effective alternative for analyzing genetic variations, particularly for SNP genotyping and methylation profiling. While high-throughput sequencing gains prominence, microarrays continue to be widely employed in large-scale genomic studies due to their affordability and efficiency.
Statistical Methods and Computational Tools
The success of genome-wide analysis depends heavily on advanced statistical methods and computational tools. Techniques such as multi-trait analysis, polygenic risk scoring, and machine learning enable researchers to interpret complex genomic datasets. Bioinformatics pipelines enhance data processing, ensuring that subtle genetic effects are accurately identified and understood.
Conclusion
Genome-wide analysis has redefined genetic research, offering transformative insights into the genetic architecture of traits and diseases. By integrating methodologies like GWAS, methylation studies, and transcriptomics with advanced technologies and computational tools, this field drives innovation in personalized medicine, agriculture, and disease prevention. As research continues to evolve, genome-wide analysis promises to unlock new frontiers in genetics and genomics, shaping the future of science and society.
References:
- Alseekh, S., Kostova, D., Bulut, M., & Fernie, A. R. (2021). Genome-wide association studies: assessing trait characteristics in model and crop plants. Cellular and molecular life sciences : CMLS, 78(15), 5743–5754. https://doi.org/10.1007/s00018-021-03868-w
- Vegt, R., Bertoli-Avella, A., Tulen, J. et al. Genome-wide linkage analysis in a Dutch multigenerational family with attention deficit hyperactivity disorder. Eur J Hum Genet 18, 206–211 (2010). https://doi.org/10.1038/ejhg.2009.148
- Habibi, E., Brinkman, A. B.,et.al. (2013). Whole-genome bisulfite sequencing of two distinct interconvertible DNA methylomes of mouse embryonic stem cells. Cell stem cell, 13(3), 360–369. https://doi.org/10.1016/j.stem.2013.06.002
- Wang, C., Lin, J., et.al. (2024). Genome-wide transcriptome analysis reveals key regulatory networks and genes involved in the determination of seed hardness in vegetable soybean. Horticulture research, 11(5), uhae084. https://doi.org/10.1093/hr/uhae084


 Sample Submission Guidelines
Sample Submission Guidelines
 Fig. 1. A schematic view of GWAS in plants.(Alseekh, et.al, 2021).
Fig. 1. A schematic view of GWAS in plants.(Alseekh, et.al, 2021). Fig. 2. Family tree showing microsatellite haplotypes.(Vegt, et.al, 2010)
Fig. 2. Family tree showing microsatellite haplotypes.(Vegt, et.al, 2010) Fig. 3.Comparison of methylation modification of mouse embryonic stem cells in different states.(Habibi, et.al, 2013)
Fig. 3.Comparison of methylation modification of mouse embryonic stem cells in different states.(Habibi, et.al, 2013) Fig. 4. Differentially expression genes in different samples.(Wang,et.al,2024)
Fig. 4. Differentially expression genes in different samples.(Wang,et.al,2024)