Revolutionizing MSI Detection with Next-Generation Sequencing (NGS)
When we talk about genetic testing and cancer diagnostics, Next-Generation Sequencing (NGS) stands at the forefront of innovation. The ability to examine an entire genome in minute detail has completely shifted the way we think about Microsatellite Instability (MSI)—a key indicator of genetic instability that plays a pivotal role in the development of various cancers. Unlike traditional methods like PCR or immunohistochemistry (IHC), which often focus on just a few microsatellite markers, NGS dives deeper, providing a holistic view of microsatellite regions across the entire genome.
This comprehensive approach offers much more than just a "yes" or "no" on whether MSI is present; it allows clinicians to classify tumors into distinct categories: MSI-high, MSI-low, and microsatellite stable (MSS). This classification is crucial because it doesn't just reveal the presence of genomic instability, but it also paves the way for more personalized, targeted treatment decisions—especially in the realm of immunotherapy.
The article dives deep into the practical applications of MSI in population genetics, with a special focus on how NGS technology advances MSI detection. It offers a comprehensive overview of MSI, from its definition and underlying genetic mechanisms to its diagnostic value. NGS doesn't just improve the sensitivity and accuracy of MSI detection; it enhances our ability to interpret its implications, particularly in cancer treatment. The piece also explores the clinical significance of MSI, especially in cancer diagnosis and prognosis, as well as detailing the process of MSI testing through NGS. Industry trends, current challenges, and promising future developments in MSI testing are also addressed, providing readers with a well-rounded understanding of this exciting area of genomic medicine.
Microsatellites and Their Role in Genome Stability
What Are Microsatellites?
Microsatellites are short, repetitive sequences of DNA that are prone to errors during DNA replication. These regions are highly polymorphic, meaning their length can vary significantly between individuals. Although microsatellites make up a small portion of the genome, their repetitive nature plays a significant role in maintaining genome stability.
Mechanisms of Microsatellite Instability
MSI occurs when the DNA mismatch repair system fails to correct errors during DNA replication, leading to insertions or deletions in microsatellite regions. Over time, this results in a hypermutable phenotype, where the number of mutations in key genes increases, contributing to cancer progression. The genetic pathways involved in MSI are complex but often include defects in MMR proteins such as MLH1, MSH2, and PMS2.
NGS and Its Impact on MSI Testing
- Comprehensive Analysis: WGS offers a comprehensive approach by analyzing the entire genome, identifying MSI across both known and previously uncharacterized regions. This method uncovers MSI events that may be missed by targeted approaches.
- Increased Detection Sensitivity: WGS increases the likelihood of detecting MSI events throughout the genome, providing a more complete view of genomic instability in cancers. For example, Aaltonen et al. (2020) demonstrated that WGS identified MSI in tumors that traditional methods failed to detect, particularly in rare microsatellite regions.
- Cost-Effective: Targeted panel sequencing focuses on predefined microsatellite regions, offering a cost-effective alternative to WGS. This method is highly sensitive and accurate, particularly in clinical settings where MSI is evaluated in well-characterized regions.
- Clinical Validation: Multiple studies, including Liu et al. (2019), have shown that targeted panels can reliably detect MSI, providing accuracy comparable to WGS, but at a reduced cost, making it a practical option for routine clinical use.
Comparison with Traditional MSI Testing Methods
1. Polymerase Chain Reaction (PCR)
- Limitations: PCR is limited in its ability to analyze large genomes or tumors with complex MSI patterns. It often fails to detect MSI in certain microsatellite regions, particularly those with high genomic instability (Liu et al., 2020).
- Reduced Scope: PCR typically analyzes a limited number of loci (5-7), making it less reliable for capturing the full spectrum of MSI, especially in heterogeneous tumors.
2. Immunohistochemistry (IHC)
- Indirect Assessment: IHC detects mismatch repair (MMR) protein expression, but it does not directly assess MSI at the molecular level. While useful for identifying loss of MMR protein expression, it can miss MSI events that are not associated with MMR protein loss.
- Inconsistencies: MSI-high tumors may exhibit MMR protein expression loss in some areas but not in others, complicating IHC-based assessments (Kruiswijk et al., 2021). Hence, IHC offers less precise information compared to molecular techniques like NGS.
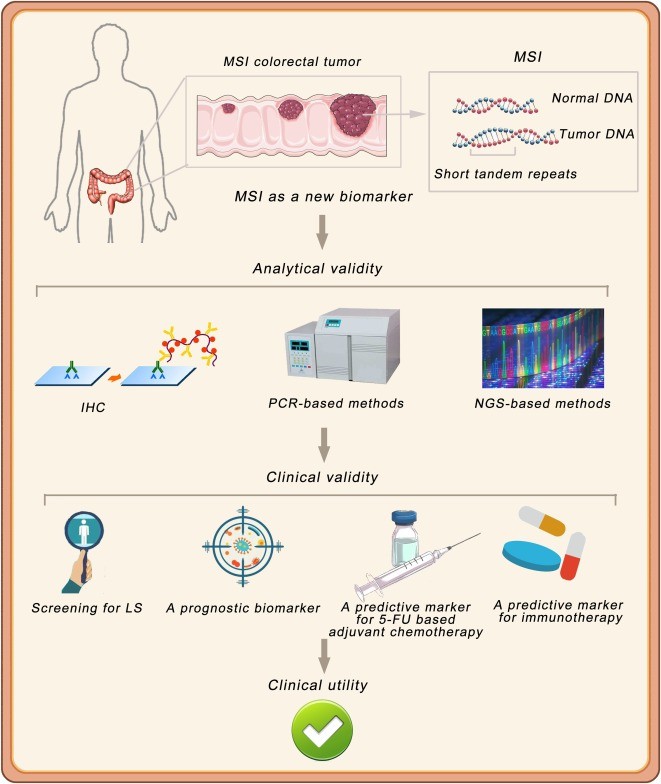 MSI Detected by different methods
MSI Detected by different methods
Advantages of NGS Over Traditional Methods
1. Comprehensive Analysis
- Genome-Wide Assessment: NGS analyzes thousands of microsatellite loci across the entire genome, whereas PCR-based methods only analyze a few loci (typically 5-7). This provides a much more thorough assessment of MSI.
2. Improved Performance
- Higher Sensitivity and Specificity: NGS methods demonstrate higher sensitivity and specificity than traditional techniques. Some NGS approaches have shown sensitivity of 97.0% (95% CI, 89.6%-99.6%) and specificity with a positive predictive value > 95.0% (Xie et al., 2021), outperforming PCR and IHC in terms of accuracy.
3. Limitations of Traditional Methods
- Cost and Labor-Intensive: PCR is often costly and labor-intensive, and may miss the full spectrum of MSI, particularly in large genomes (Smith et al., 2018).
- Indirect MSI Detection in IHC: While IHC detects MMR protein expression, it does not directly assess MSI, making it less precise than NGS.
4. High Concordance with Traditional Methods
- Accuracy in Colorectal Cancers: NGS methods have demonstrated high concordance with PCR and IHC assessments, especially in colorectal cancers with tumor cell percentages ≥ 30%. Some studies found 100% sensitivity and specificity for NGS compared to IHC (Zhang et al., 2019).
5. Additional Advantages of NGS
- Simultaneous Genomic Insights: NGS allows for the simultaneous analysis of MSI status and other genomic alterations in a single test, providing a more comprehensive understanding of the tumor's genetic profile (Huang et al., 2021).
- No Need for Matched Normal Tissue: Some NGS methods can determine MSI status without requiring matched normal tissue samples, making the testing process more convenient and less invasive (Li et al., 2020).
While traditional methods like PCR and IHC continue to play a role in MSI testing, NGS offers a more comprehensive, sensitive, and specific approach to detecting MSI in cancer. By analyzing the entire genome, NGS increases the accuracy of MSI detection, improving diagnostic outcomes and offering greater insight into cancer genomics. As NGS technology evolves, it is expected to become the gold standard for MSI testing, providing essential tools for personalized cancer treatment.
Practical Steps in MSI Testing Using NGS
Sample Collection and Preparation
To evaluate MSI, tumor samples are typically collected either from tissue biopsies or, increasingly, from liquid biopsies (such as blood or other bodily fluids). Liquid biopsies provide a less invasive option for obtaining genetic information and are being used more frequently in clinical settings for MSI detection. The ease and convenience of liquid biopsies make them an appealing choice for real-world applications, offering patients a less disruptive alternative to traditional tissue biopsies.
NGS Data Processing for MSI Analysis
Once the tumor samples are collected, NGS is employed to process the DNA. The sequencing reads are first aligned to a reference genome, ensuring that the data is mapped accurately to known genomic regions. The next step is identifying mutations within the microsatellite regions, which are key to detecting MSI. By comparing these mutations to normal tissue samples or population-based reference data, bioinformatics tools can classify the MSI status of the tumor.
Advanced bioinformatics pipelines are used to analyze the data, taking into account various factors such as insertion/deletion (indel) patterns and tumor content. Specialized algorithms like MSIdetect or MSIPeak are applied to interpret the instability in microsatellite loci, further refining the MSI classification process.
Interpreting MSI Results
After the data has been processed, the MSI results are classified into one of three categories:
MSI-high (MSI-H): This classification indicates a high number of mutations in the microsatellite regions, which can significantly impact treatment decisions, particularly in the context of immunotherapy. MSI-high tumors are often more responsive to immune checkpoint inhibitors, making this classification crucial for personalized cancer treatment.
MSI-low (MSI-L): Tumors in this category exhibit moderate instability, with fewer mutations in the microsatellite regions compared to MSI-high tumors. This can influence treatment options, though to a lesser degree than MSI-high.
Microsatellite Stable (MSS): Tumors in the MSS category show no detectable instability in the microsatellite regions, suggesting a stable genomic profile. MSI testing is often used to rule out MSI in cases where other treatment avenues, such as immunotherapy, are being considered.
This classification of MSI status is crucial for guiding clinical decisions and tailoring treatment strategies. NGS-based MSI testing offers an enhanced, detailed view of the tumor's genomic landscape, allowing for more accurate, personalized treatment plans, especially in the rapidly evolving field of cancer immunotherapy.
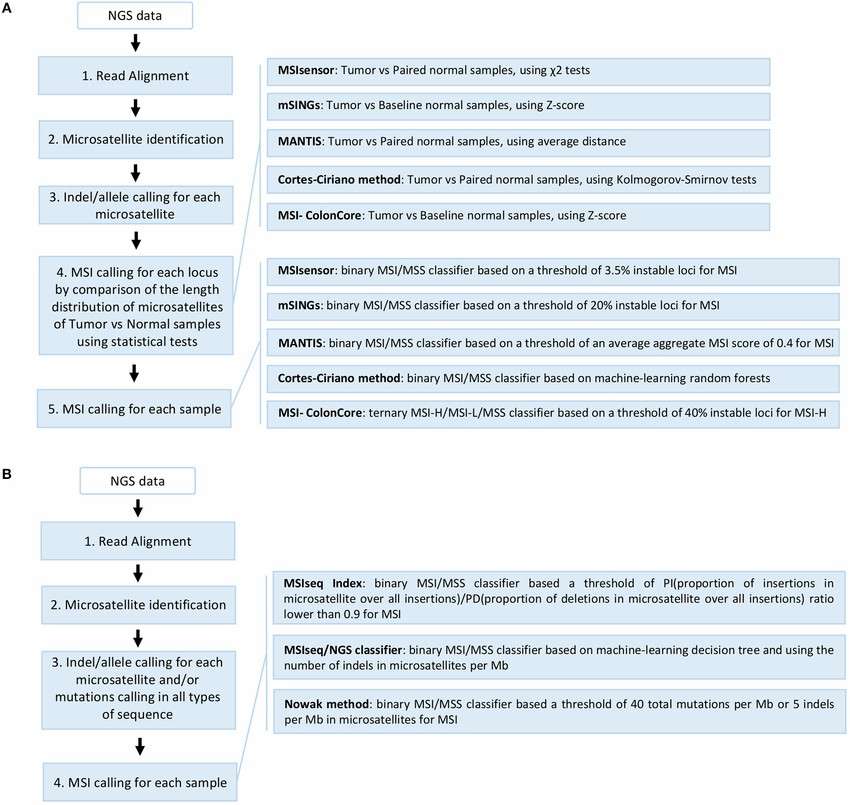 Overview of the different NGS-based computational methods developed for MSI detection (Laura G. Baudrin et al,. 2018)
Overview of the different NGS-based computational methods developed for MSI detection (Laura G. Baudrin et al,. 2018)
Industry Trends in MSI Testing and NGS
FDA-Approved NGS Panels for MSI Testing
Several FDA-approved NGS panels now exist for MSI testing, making it easier for clinicians to integrate MSI testing into routine clinical practice. These panels include targeted assays designed specifically to evaluate microsatellite regions and identify MSI status.
Liquid Biopsy and MSI Detection
Liquid biopsy is gaining momentum as a non-invasive alternative for MSI testing. By analyzing cfDNA (circulating free DNA) in blood samples, clinicians can monitor MSI status without the need for traditional tissue biopsies. Liquid biopsy is especially valuable for early cancer detection and monitoring treatment response.
Challenges and Limitations of NGS in MSI Testing
Technical Challenges
While NGS is a powerful tool, several technical challenges exist. Issues like sequencing depth, sample quality, and cost can limit the widespread adoption of NGS in routine clinical settings. Ensuring high-quality DNA samples and adequate sequencing depth is essential for reliable MSI detection.
Clinical Challenges
Clinically, NGS testing for MSI faces challenges related to sensitivity and specificity, particularly in certain cancer types where MSI might be subtle or in low-frequency cases. Proper validation and standardization are necessary to improve the reliability of MSI detection.
Future Directions and Innovations in MSI Testing and NGS
Artificial Intelligence and Machine Learning in MSI Detection
Advancements in artificial intelligence (AI) and machine learning (ML) are poised to enhance MSI detection. By analyzing large datasets, AI could potentially identify subtle patterns in MSI that are difficult for human analysis, improving the accuracy and speed of diagnostics.
Advancements in NGS Platforms
The continuous evolution of NGS technology, with platforms like PacBio and Oxford Nanopore, promises to make MSI testing even more precise. These technologies offer longer read lengths and higher accuracy, which could be crucial for detecting MSI in more complex genomes.
Conclusion
In summary, Microsatellite Instability (MSI) is a crucial factor in cancer progression, and Next-Generation Sequencing (NGS) has become the gold standard for detecting MSI with high sensitivity and accuracy. Understanding MSI's role in various cancers, including colorectal and endometrial cancers, is essential for early diagnosis and personalized treatment planning, particularly in the context of immunotherapy.
At CD Genomics, we are dedicated to providing advanced microsatellite genotyping and MSI analysis services to help researchers advance their understanding of cancer genomics. Learn more about our cutting-edge services and explore our full range of genomic solutions:
References:
-
Boland, C. R., & Goel, A. (2010). Microsatellite instability in colorectal cancer. Gastroenterology, 138(6), 2073-2087.e3. https://doi.org/10.1053/j.gastro.2009.12.064
- Le, D. T., Durham, J. N., Smith, K. N., Wang, H., Bartlett, B. R., Aulakh, L. K., ... & Diaz, L. A. (2017). Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science, 357(6349), 409-413. https://doi.org/10.1126/science.aan6733
- Sinicrope, F. A. (2019). Lynch syndrome–associated colorectal cancer. New England Journal of Medicine, 379(8), 764-773. https://doi.org/10.1056/NEJMcp1714533
- Vilar, E., & Gruber, S. B. (2010). Microsatellite instability in colorectal cancer—the stable evidence. Nature Reviews Clinical Oncology, 7(3), 153-162. https://doi.org/10.1038/nrclinonc.2009.237
- Cohen, R., Buhard, O., Cervera, P., Hain, E., Dumont, S., Bardier, A., ... & André, T. (2017). Clinical and molecular characterisation of hereditary and sporadic metastatic colorectal cancers harbouring microsatellite instability/DNA mismatch repair deficiency. European Journal of Cancer, 86, 266-274. https://doi.org/10.1016/j.ejca.2017.09.022


 Sample Submission Guidelines
Sample Submission Guidelines
 MSI Detected by different methods
MSI Detected by different methods  Overview of the different NGS-based computational methods developed for MSI detection (Laura G. Baudrin et al,. 2018)
Overview of the different NGS-based computational methods developed for MSI detection (Laura G. Baudrin et al,. 2018) 