What is Microsatellite Instability
Microsatellite instability (MSI) is a phenomenon characterized by alterations in the length of microsatellites, which are repetitive DNA sequences dispersed throughout the genome. These microsatellites, also referred to as simple sequence repeats (SSRs) or short tandem repeats (STRs), typically consist of repeat units spanning 1-6 nucleotides. In humans, microsatellites predominantly reside in non-coding regions, although they can also be present in coding regions.
During DNA replication, microsatellites are particularly susceptible to slippage events, wherein the DNA polymerase may insert or delete nucleotide repeat units, leading to changes in their length. Normally, these alterations are corrected by the mismatch repair (MMR) system. However, when components of the MMR system are defective—due to mutations or epigenetic modifications—microsatellite instability arises.
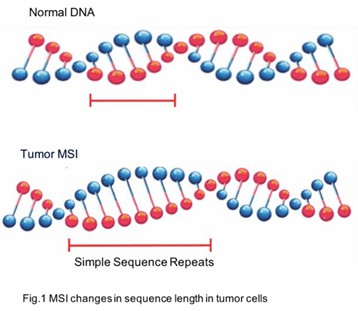
What is a Microsatellite Instability Test
A MSI test is a diagnostic procedure designed to identify the presence of MSI within a DNA sample. This test is pivotal for elucidating the genetic underpinnings of certain cancers and for tailoring appropriate therapeutic strategies. MSI tests typically detect discrepancies in the length of microsatellite repeats, which can signify defects in the MMR system.
Various methodologies are employed for MSI testing, each with distinct applications and sensitivities. These tests are particularly crucial in cancer diagnostics, notably for colorectal cancers and Lynch syndrome, where high levels of MSI can indicate a genetic predisposition to cancer.
Methods of Microsatellite Instability Test
- PCR with Capillary Electrophoresis
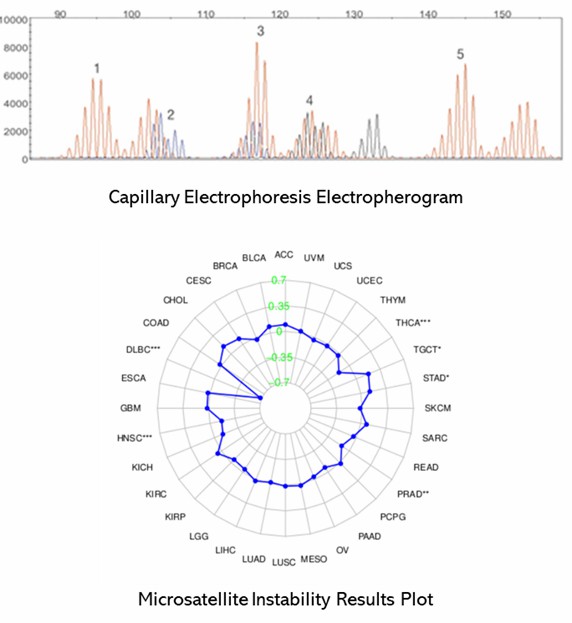
The conventional gold standard for MSI testing combines polymerase chain reaction (PCR) with capillary electrophoresis. This approach begins with the amplification of microsatellite regions using fluorescently labeled primers. Subsequently, the amplified products are separated by size through capillary electrophoresis, which allows for the detection and analysis of variations in repeat lengths.
The process starts with the extraction of DNA from both tumor and normal tissue samples. Specific microsatellite loci are then amplified via PCR, with one primer labeled with a fluorescent dye. The PCR products are subjected to capillary electrophoresis, which separates them based on size and enables the detection of MSI by analyzing the differences in repeat lengths. This method is highly regarded for its specificity and reliability in detecting MSI, maintaining its status as the gold standard.
- PCR with High-Resolution Melt Curve Analysis
Another method for MSI testing involves PCR coupled with high-resolution melt (HRM) curve analysis. This technique uses DNA intercalating dyes to monitor changes in the melting temperature (Tm) of DNA fragments, which provides insights into microsatellite stability.
The procedure involves amplifying DNA fragments in the presence of a high-resolution dye. After amplification, the DNA is gradually heated while monitoring the fluorescence signal to detect differences in melting temperatures. HRM analysis is particularly sensitive to minor length variations and is effective in identifying MSI, including single-base changes and small insertions or deletions.
- Next-Generation Sequencing (NGS)
Next-Generation Sequencing (NGS) technologies provide a high-throughput method for detecting MSI by sequencing multiple microsatellite loci across the genome. This approach can be applied through whole-genome sequencing (WGS), whole-exome sequencing (WES), or targeted region sequencing (TRS) to identify MSI and other genetic variations.
The process involves using high-throughput sequencing technologies to sequence microsatellite regions. The sequencing data are then analyzed to detect variations in microsatellite repeat lengths and identify MSI. NGS offers a comprehensive view of microsatellite instability along with other genetic mutations, providing an in-depth genetic profile of the tumor.
Introduction of Microsatellite Instability Analysis Service
CD Genomics employs Promega kit which is a fluorescent multiplex PCR-Capillary Electrophoresis based method to detect the MSI status. Typically, the MSI analysis system includes allelic profiles of microsatellite markers comparison, involving five nearly monomorphic mononucleotide repeat markers (BAT-25, BAT-26, MONO-27, NR-21 and NR-24) and two highly polymorphic pentanucleotide repeat markers (Penta C and Penta D), which are quality control for sample authentication of matched normal and tumor. For MSI detection, tumor and normal tissues from each patient need to be analyzed in parallel. Generally, MSI detection in ≥ 30-40% of the markers is considered to be MSI-high (MSI-H), whereas that in < 30-40% of the markers is considered MSI-low (MSI-L), or MSS if no marker is unstable.
Advantages and Applications of Microsatellite Instability Analysis
- Greater accuracy than IHC, cost-effective and faster turnaround than NGS
- Lynch syndrome screening
- Prognosis detection in sporadic colorectal cancer
- Diagnostic tests in a wide range of solid tumors
Microsatellite Instability Analysis Workflow

Service Specifications
| Sample Requirements 1) For normal tissue
|
|
Click |
Test Strategy
|
| Bioinformatics Analysis We provide multiple customized bioinformatics analyses:
|
Analysis Pipeline
Deliverables
- Raw data
- Experimental results
- Data analysis report
- Details in Microsatellite Instability Analysis for your writing (customization)
CD Genomics offers an advanced microsatellite instability analysis service leveraging state-of-the-art technologies such as PCR, HRM, and NGS. This service is designed to meet the needs of researchers seeking detailed insights into microsatellite instability in various cancer types. By employing cutting-edge methodologies and high-throughput technologies, CD Genomics ensures accurate and reliable MSI detection.
References:
- Le, D.T. et al. (2017) Mismatch-repair deficiency predicts response of solid tumors to PD-1 blockade. Science 10.1126/scienc.aan6733.
- Le, D.T. et al. (2015) PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. New Engl. J. Med. 372, 2509–20.
- NCCN Clinical Practice Guidelines in Oncology: Genetic/Familial High-Risk Assessment: Colorectal. Version 2.2015.
- Promega Technical Manual. MSI Analysis System v1.2. Oct 2012.
Partial results are shown below:
1. What is the significance of MSI in cancer diagnosis?
Microsatellite instability (MSI) serves as a crucial marker indicative of defective mismatch repair systems and is closely associated with various cancers, particularly those linked to Lynch syndrome. The detection of MSI provides essential information across multiple dimensions, including diagnosis, prognosis, and treatment planning. By identifying MSI, clinicians can better characterize the genetic underpinnings of cancer, leading to more precise and informed medical decisions.
2. How does NGS improve MSI testing?
NGS significantly advances MSI testing by enabling high-throughput, comprehensive analysis of multiple microsatellite loci. This technology offers detailed insights into both MSI and other genetic variations, thus enhancing the accuracy and depth of genetic analysis. NGS's ability to simultaneously evaluate numerous loci allows for a more holistic understanding of the genome, leading to improved diagnostic precision and potentially uncovering novel therapeutic targets.
3. Can MSI testing be performed on non-tumor samples?
MSI testing is not confined to tumor tissue samples; it can also be conducted on various other sample types, including blood samples through the analysis of circulating tumor DNA (ctDNA). While tumor tissue samples are generally preferred for their accuracy, the capability to use non-invasive samples expands the practicality of MSI testing in clinical settings, particularly for monitoring disease progression and treatment response.
4. What are the benefits of using CD Genomics' MSI analysis service?
CD Genomics offers an advanced and high-sensitivity MSI testing service that leverages the latest in genomic technologies. The company provides tailored solutions designed to meet the specific needs of both research and clinical application. Expert support ensures the delivery of high-quality results and comprehensive data, making CD Genomics a reliable partner for sophisticated MSI analysis. Their commitment to state-of-the-art methodologies ensures that clients receive the most accurate and actionable insights for their projects.
Low temperature isothermal amplification of microsatellites drastically reduces stutter artifact formation and improves microsatellite instability detection in cancer
Journal: Nucleic acids research
Impact factor: 16.6
Published: 17 September 2019
Background
Microsatellites are repetitive DNA sequences used in genetic research and cancer diagnostics. MSI, an alteration in cancer due to DNA repair defects, is typically detected via PCR and capillary electrophoresis. The authors introduced low-temperature isothermal amplification with recombinase polymerase amplification (RPA) as a promising alternative, reducing PCR artifacts and improving MSI detection and allele calling.
Materials & Methods
Sample Preparation:
- Human blood samples
- Patient colorectal cancer samples
- DNA extraction
Method:
- Polymerase chain reaction (PCR)
- Recombinase polymerase amplification (RPA)
- Genotyping
- DNA amplicon sequencing
- Capillary electrophoresis fragment analysis
- Fragment length analysis
- Microsatellite allele read counts
Results
In evaluating mono- to tetra-nucleotide repeat microsatellite LT-RPA assays on blood samples, LT-RPA demonstrated a significant improvement over PCR by simplifying microsatellite profiles through the reduction or elimination of stutter peaks, thereby making allele identification clearer and more accurate. This was particularly evident for challenging microsatellites like BAT26, D18S61, and D12ATA63, where PCR often resulted in stutter peaks and amplification biases that complicated genotype calling. Amplicon sequencing further confirmed that LT-RPA profiles were consistent with PCR results, except for a specific polymorphism, and allowed precise allele identification.
When assessing tri-nucleotide repeat microsatellites implicated in genetic diseases, LT-RPA effectively amplified CTG and GAA repeats with reduced stutter peaks compared to PCR, although it struggled with CG-rich FMR1 microsatellites. For HTT CAG repeats, LT-RPA provided improved results with optimized reaction conditions, demonstrating its capability in handling complex repeat structures.
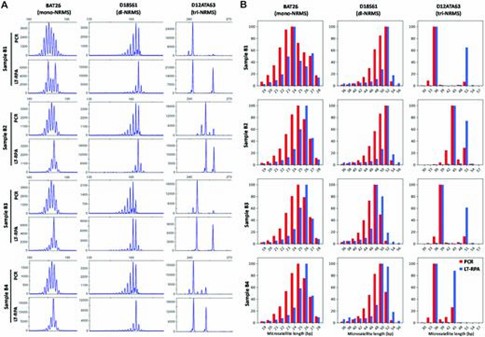 Figure 1. Microsatellite profiles of BAT26, D18S61 and D12ATA63 obtained with four blood samples (named B1, B2, B3 and B4) from healthy individuals using PCR (20 ng of DNA in 20 μl) and LT-RPA.
Figure 1. Microsatellite profiles of BAT26, D18S61 and D12ATA63 obtained with four blood samples (named B1, B2, B3 and B4) from healthy individuals using PCR (20 ng of DNA in 20 μl) and LT-RPA.
In testing HT17 LT-RPA on serial dilutions of cancer cell lines, LT-RPA improved the limit of detection (LOD) for microsatellite instability (MSI), especially for smaller deletions, by minimizing stutter peaks that hindered detection in PCR. LT-RPA also facilitated better identification of mutant alleles in mixed samples, addressing the issues of stutter peaks present in PCR.
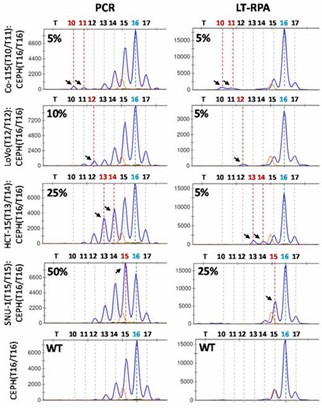 Figure 2. HT17 LT-RPA improves the limit of detection of MSI compared to PCR.
Figure 2. HT17 LT-RPA improves the limit of detection of MSI compared to PCR.
Finally, LT-RPA proved to be highly effective in detecting MSI in CRC samples, including those where PCR failed due to stutter peaks. The method provided better clarity and accuracy, and validation with small-pool PCR confirmed LT-RPA results and detected additional mutated alleles, although some findings might be artifacts from PCR.
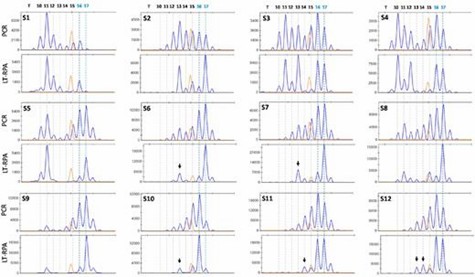 Figure 3. Detection of MSI in 12 fresh frozen MMR-deficient CRC samples (S1-S12) using HT17 microsatellite and LT-RPA and PCR.
Figure 3. Detection of MSI in 12 fresh frozen MMR-deficient CRC samples (S1-S12) using HT17 microsatellite and LT-RPA and PCR.
Conclusion
The authors developed a low-temperature RPA method for microsatellites that reduces stutter artifacts and simplifies allele calling. This method improves MSI detection in colorectal cancer and shows strong potential as a PCR alternative for both clinical and research use.
Reference:
- Daunay A, Duval A, Baudrin LG, et al. Low temperature isothermal amplification of microsatellites drastically reduces stutter artifact formation and improves microsatellite instability detection in cancer. Nucleic acids research. 2019, 47(21):e141-.
Here are some publications that have been successfully published using our services or other related services:
Fungi: friends or foes—an outreach science initiative for the collection of airborne fungal spores by high school students
Journal: Journal of Microbiology and Biology Education
Year: 2024
Small but significant genetic differentiation among populations of Phyllachora maydis in the midwestern United States revealed by microsatellite (SSR) markers
Journal: bioRxiv
Year: 2023
The genetic legacy of fragmentation and overexploitation in the threatened medicinal African pepper-bark tree, Warburgia salutaris
Journal: Scientific Reports
Year: 2020
Evaluation of Plasma Biomarkers for A/T/N Classification of Alzheimer Disease Among Adults of Caribbean Hispanic Ethnicity
Journal: JAMA Network Open
Year: 2023
See more articles published by our clients.


 Sample Submission Guidelines
Sample Submission Guidelines
