1. Introduction: What Are Microsatellite Markers?
What Are Microsatellites?
Microsatellites, also known as simple sequence repeats (SSRs), are short, repetitive DNA sequences consisting of 1–6 base pairs. These sequences are highly polymorphic, meaning they exhibit significant variation in length between individuals, making them valuable tools in a wide range of genetic research. Found throughout the genome, microsatellites are particularly useful in studies such as genetic mapping, population genetics, parentage testing, and even forensic science.
Why Microsatellite Markers Matter in Genomics
Microsatellite markers are prized in genomic studies for their high mutation rate and co-dominant inheritance, which allows researchers to distinguish between two different alleles at a given locus. Their widespread occurrence across species also means they are useful in genetic diversity studies, marker-assisted selection, and evolutionary biology. Whether for agriculture, forensics, or conservation genetics, these markers are indispensable in modern genetics.
2. What Is a Microsatellite Marker Database?
Definition and Role in Genetic Research
A microsatellite marker database is an organized collection of data that stores information about microsatellites across various organisms. These databases are essential in genomic research as they provide researchers with accurate, accessible, and comprehensive information about microsatellites, their locations, and their role in different species. By compiling this data in a central place, they allow scientists to accelerate research, improve marker identification, and enhance genetic mapping efforts.
Key Features of a Microsatellite Marker Database
Effective microsatellite marker databases have several key features:
- Extensive coverage of species and genetic markers.
- Ease of access via open or subscription-based platforms.
- Quality control to ensure the accuracy and consistency of data.
- Regular updates to accommodate the evolving body of genetic knowledge. These features ensure that researchers can rely on these databases for high-quality, relevant data.
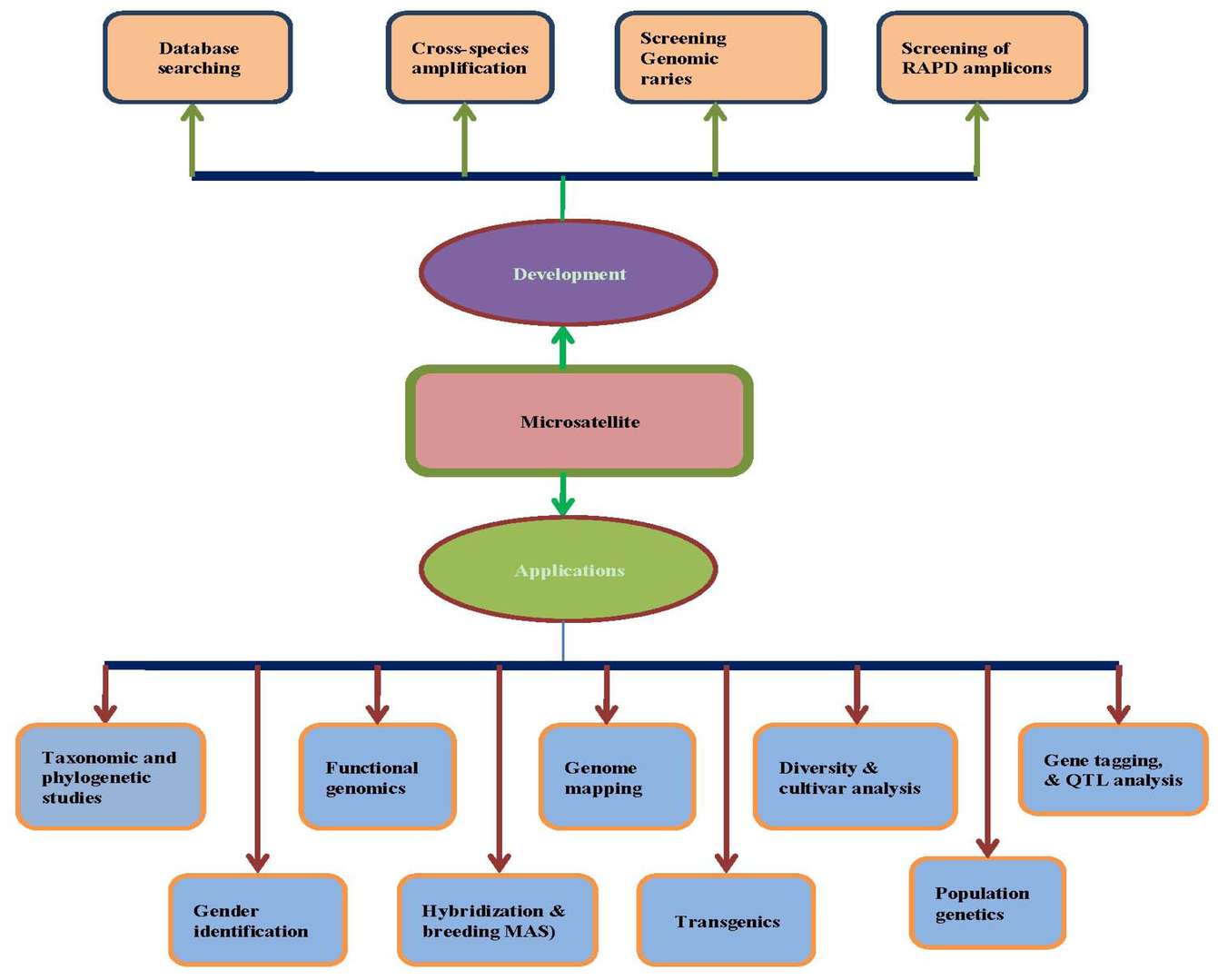 Figure 1. Development and applications of microsatellite markers at a glance (Gous Miah et al,. 2013)
Figure 1. Development and applications of microsatellite markers at a glance (Gous Miah et al,. 2013)
3. Popular Microsatellite Marker Databases: An Overview
Comprehensive Databases
| Database Name |
Description |
Key Features |
| MSDB (MicroSatellite DataBase) |
Contains over 4 billion microsatellites from 37,680 genomes, one of the largest resources for researchers. |
Large-scale collection, extensive genome coverage, interactive portal. |
| MICdb3.0 |
Updated version of MICdb, focusing on perfect microsatellites from completely sequenced genomes of bacteria and archaea. |
Specializes in microbial genomes, well-annotated sequences. |
| pSATdb |
Provides data on polymorphic mitochondrial microsatellites across 5,976 mitochondrial genome sequences from 1,576 genera. |
Focus on mitochondrial SSRs, rich in polymorphic data. |
| SSRome |
A large-scale resource with 45.1 million microsatellite markers across various taxa, including plants, metazoans, and others. |
Massive dataset, includes a wide variety of organisms. |
Specialized Databases
| Database Name |
Description |
Key Features |
| LegumeSSRdb |
Contains 3,706,276 SSRs from 13 legume species, including both genic and non-genic SSRs. |
Focus on legumes, provides both genic and non-genic data. |
| Cotton Microsatellite Database |
Catalogs 5,484 SSR markers derived from nine major cotton microsatellite projects. |
Specialized for cotton research, marker-based crop studies. |
| Kazusa Marker Database |
Provides linkage and physical maps, and data on approximately 68,000 SSR primers for 14 agronomically important crops. |
Focus on crop improvement, large SSR catalog. |
| PIPEMicroDB |
Contains 123,387 short tandem repeats identified in the pigeon pea genome. |
Specialized for pigeon pea, supports crop breeding research. |
| FmMDb (Foxtail Millet Marker Database) |
Catalogs 21,315 genomic SSRs, 447 genic SSRs, and 96 intron length polymorphisms. |
Focus on foxtail millet, valuable for genomic studies. |
| CicArMiSatDB (Chickpea Microsatellite Database) |
Derived from chickpea genome data, provides extensive SSR information for chickpeas |
|
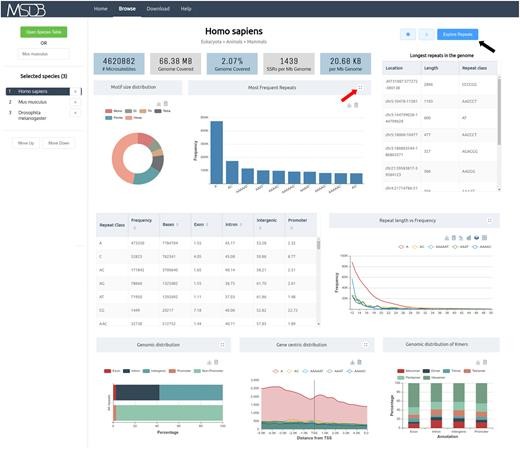 Figure 2. Browse page of MSDB showing microsatellite information (Akshay Kumar Avvaru et al,.2020)
Figure 2. Browse page of MSDB showing microsatellite information (Akshay Kumar Avvaru et al,.2020)
Comparing Microsatellite Marker Databases
| Database Name |
Number of Species |
Number of Microsatellites |
Major Applications |
Access Type |
| dbSNP |
50+ |
10 million+ |
SNPs, microsatellites for multiple species |
Open Access |
| GenBank |
100+ |
5 million+ |
Genomic sequences, microsatellites |
Open Access |
| MSTR (Plant) |
30+ |
500,000+ |
Plant breeding, genetic mapping |
Open Access |
| SSR Map |
60+ |
2 million+ |
Crop research, marker-assisted selection |
Open Access |
| Micromap |
10 |
100,000+ |
Forensic science, population genetics |
Subscription |
| MSDB (MicroSatellite DataBase) |
37,680+ |
4 billion+ |
Large-scale genome data, microsatellite identification |
Open Access |
| MICdb3.0 |
Bacteria/Archaea |
N/A |
Microbial genomics, perfect microsatellites |
Open Access |
| pSATdb |
1,576 genera |
N/A |
Mitochondrial genome research, polymorphic SSRs |
Open Access |
| SSRome |
Multiple taxa |
45.1 million+ |
Large-scale cross-taxa microsatellite data |
Open Access |
| LegumeSSRdb |
13 |
3,706,276 |
Legume research, SSR data for breeding |
Open Access |
| Cotton Microsatellite Database |
9 |
5,484 |
Cotton breeding, crop improvement |
Open Access |
| Kazusa Marker Database |
14 |
68,000 |
Crop genetics, agronomy research |
Open Access |
| PIPEMicroDB |
1 |
123,387 |
Pigeon pea genome research |
Open Access |
| FmMDb (Foxtail Millet Marker Database) |
1 |
21,315 |
Millet breeding, genomic SSR data |
Open Access |
| CicArMiSatDB (Chickpea Microsatellite Database) |
1 |
N/A |
Chickpea genetic studies |
Open Access |
These databases offer various features such as user-friendly interfaces, visualization tools, and the ability to export data for further analysis. They cater to diverse research needs, from plant breeding and crop improvement to forensic science and population genetics.The development of these comprehensive and specialized databases reflects the growing importance of microsatellite markers in genomics studies. Researchers can now access a wealth of information on common, polymorphic, and unique microsatellites, facilitating studies in genetic diversity, mapping, marker-assisted selection, and comparative population analyses across various species and genera.
Summary of read2Marker.
4. Applications of Microsatellite Marker Databases
In Genetic Mapping and Linkage Analysis
Microsatellite markers are integral to genetic mapping and linkage analysis, particularly in species with complex genomes. By using databases to access data on microsatellites, researchers can identify key genetic traits, improve disease resistance, and map quantitative trait loci (QTLs) for targeted breeding programs.
In Biodiversity, Conservation Genetics, and Forensic Science
Microsatellite marker databases are widely used in the study of biodiversity and conservation genetics. For instance, they help track genetic variations within endangered species and populations. In forensic science, databases such as Micromap support DNA profiling for human identification and crime scene investigation.
In Crop and Livestock Improvement
In agricultural genomics, microsatellites play a crucial role in marker-assisted selection (MAS) for improving crop yields, disease resistance, and livestock traits. Researchers can use databases to select desirable traits and accelerate the breeding process.
5. How Microsatellite Marker Databases Facilitate Genetic Research
Data Accessibility and Integration
Microsatellite marker databases provide quick access to critical genetic data across species. The ability to easily integrate data from multiple sources, such as GenBank or MSTR, helps researchers streamline their work, reducing the need for extensive manual searches and data entry.
Large-Scale Studies Utilizing Microsatellite Markers
Research projects such as genomic surveys and population studies leverage databases for large-scale analyses. For instance, next-generation sequencing (NGS) technologies use data from these databases to identify new microsatellite markers efficiently. You can explore services like microsatellite genotyping at CD Genomics to get an insight into real-world applications in large-scale studies.
6. Data Quality and Standardization in Microsatellite Marker Databases
Curating, Verifying, and Standardizing Data Across Platforms
Maintaining high-quality data in microsatellite marker databases requires rigorous curation and verification. Databases must regularly update to ensure the information is current and accurate. Platforms like dbSNP and GenBank follow strict guidelines to ensure data consistency across species and applications.
Maintaining Database Integrity
Challenges in data accuracy and consistency arise when different labs submit data with varying standards. Ensuring data quality remains intact is essential for long-term use. Databases address this by enforcing standardization protocols and using peer-reviewed datasets.
7. Technological Advances Impacting Microsatellite Marker Databases
NGS and Marker Discovery
Next-Generation Sequencing (NGS) has revolutionized the way we discover and catalog microsatellite markers. This technology allows for the rapid sequencing of genomes, significantly increasing the number of markers available in databases like dbSNP or GenBank.
Automation in Microsatellite Marker Development
Automation tools have made it easier to identify and develop microsatellite markers. Platforms like HI-SSRSeq from CD Genomics leverage automated systems to rapidly sequence and catalog microsatellites, providing researchers with a quick turnaround on their microsatellite development needs.
8. Challenges in Microsatellite Marker Database Utilization
Data Completeness, Errors, and Limited Coverage
Despite the vast amount of data in microsatellite databases, challenges still exist in ensuring data completeness and error-free entries. Researchers must often deal with missing information or outdated data, especially when working with rare species or non-model organisms.
Ethical and Privacy Concerns in Genomic Data Sharing
The sharing of genomic data, especially involving human DNA, raises concerns regarding privacy and ethical issues. As genomic data becomes increasingly available in open-access databases, safeguarding individuals' privacy remains a crucial consideration.
9. Future Trends in Microsatellite Marker Databases
Evolution with Advancements in Genomic Technologies
As genomic technologies continue to advance, the scope of microsatellite marker databases will expand. Future databases are expected to provide more detailed data, faster updates, and broader species coverage.
Impact of AI and Machine Learning on Data Mining and Analysis
Artificial Intelligence (AI) and machine learning (ML) are poised to revolutionize how researchers analyze and mine data in microsatellite marker databases. AI algorithms can rapidly identify new markers and suggest genetic associations that might have been overlooked by traditional methods.
10. Conclusion: The Growing Role of Microsatellite Marker Databases in Genomics
Microsatellite marker databases are invaluable tools in modern genomics, playing a crucial role in everything from genetic research and biodiversity studies to agriculture and forensics. As technological advances continue to shape the field, these databases will only become more integral to scientific discovery.
If you are looking to leverage the power of microsatellite markers for your research or project, explore the services offered by CD Genomics, including microsatellite genotyping, instability analysis, and marker development.
Start enhancing your research today by tapping into a wealth of data, insights, and expert services.
Relevant FAQ
What is a microsatellite marker?
Answer: Microsatellites, also known as simple sequence repeats (SSRs), are repetitive, short DNA sequences (1–6 base pairs long) found throughout the genome. They are highly polymorphic, making them ideal for genetic studies such as mapping, population genetics, and marker-assisted selection.
What is the use of microsatellite markers in genetics?
Answer: Microsatellite markers are primarily used in genetic studies for their high variability. They are employed in gene mapping, assessing genetic diversity, parentage testing, forensic analysis, and investigating evolutionary relationships in species.
What are the best microsatellite marker databases?
Answer: Some of the best-known databases include:
dbSNP: A widely used database for single nucleotide polymorphisms (SNPs) but also includes microsatellites.
GenBank: A comprehensive database maintained by NCBI, with microsatellite information for numerous species.
MSTR: A specialized microsatellite marker database for plant species, especially crops.
SSR Map: For microsatellite markers in plants, often used in agricultural research.
How do microsatellite markers work?
Answer: Microsatellite markers work by identifying variations in the number of repeated DNA sequences at a given locus. These variations, known as alleles, are detected using techniques like PCR and gel electrophoresis. The number of repeats at a microsatellite locus can vary between individuals, making them highly informative for genetic analysis.
Why are microsatellite markers important in genomics?
Answer: Microsatellite markers are valuable due to their high mutation rate, co-dominant inheritance, and widespread presence across species. They are often used for genetic diversity studies, mapping traits of interest, and assessing population structure. Their application in forensic science and paternity testing is also significant.
References:
- "Microsatellites: Evolution and Applications," Nature Reviews Genetics, 2022.
- "dbSNP: A resource for genetic variation analysis," NCBI, https://www.ncbi.nlm.nih.gov/snp/.
- "GenBank: A comprehensive sequence database," NCBI, https://www.ncbi.nlm.nih.gov/genbank/.
- "Agricultural Genomics and the Role of Microsatellites in Crop Breeding," Plant Molecular Biology, 2020.
- "The Role of Microsatellite Markers in Conservation Genetics," Trends in Ecology and Evolution, 2021.


 Sample Submission Guidelines
Sample Submission Guidelines
 Figure 1. Development and applications of microsatellite markers at a glance (Gous Miah et al,. 2013)
Figure 1. Development and applications of microsatellite markers at a glance (Gous Miah et al,. 2013) Figure 2. Browse page of MSDB showing microsatellite information (Akshay Kumar Avvaru et al,.2020)
Figure 2. Browse page of MSDB showing microsatellite information (Akshay Kumar Avvaru et al,.2020)