Introduction to Genetic Markers
Genetic markers are like signposts in our DNA, helping us pinpoint specific genes, track traits, or even identify individuals or species. They're essential tools in molecular biology, forensic science, and genetic research.
When it comes to genetic markers, microsatellites and SNPs (Single Nucleotide Polymorphisms) are two of the most commonly used. But what's the difference between them? And how do you decide which one to use for your research?
Understanding these markers is key to making the right choice for your study. Let's explore how microsatellites and SNPs compare, and what makes them ideal for different research needs.
Types of Genetic Markers
Genetic markers are essentially like signposts in the genome, guiding researchers through the vast landscape of DNA. Whether it's for tracing inheritance patterns, identifying disease-related genes, or studying evolutionary relationships, these markers provide crucial insights. Let's break down some of the most common types:
1. Single Nucleotide Polymorphisms (SNPs)
SNPs, or Single Nucleotide Polymorphisms, are the reigning champions when it comes to genetic variation. They represent the smallest change in the genetic code: just a single nucleotide difference between individuals. And while that might sound subtle, these little shifts are incredibly powerful.
SNPs have some undeniable advantages:
- Abundant and evenly spread: They're like the punctuation marks scattered across the entire genome, making them perfect for high-resolution genetic studies.
- Automated for large-scale research: They're straightforward to analyze, especially in massive studies like genome-wide association studies (GWAS).
- Highly useful in understanding traits and diseases: If you're trying to link genetic factors to diseases or traits, SNPs are the go-to markers.
Their presence in everything from population genetics to personal genomics solidifies their place as one of the most widely used genetic markers in modern research.
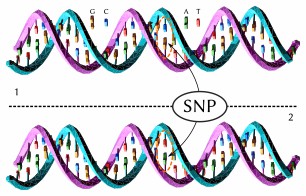 The upper DNA molecule differs from the lower DNA molecule at a single base-pair location (wikipedia)
The upper DNA molecule differs from the lower DNA molecule at a single base-pair location (wikipedia)
2. Simple Sequence Repeats (SSRs) or Microsatellites
Next up are SSRs, often called microsatellites—which are essentially repeated sequences of DNA that vary in length from one individual to another. These markers might seem simple, but they're incredibly rich in information.
Here's why microsatellites are such a big deal:
- High polymorphism: Their variability makes them fantastic for studying genetic diversity, which is why they're often used in forensic science and population studies.
- Co-dominant inheritance: This means both copies of the gene (from mom and dad) can be detected, which is great for studies in inheritance patterns.
- Powerful in breeding: If you're involved in marker-assisted selection (MAS) for improving crops or animals, microsatellites are like a trusty sidekick.
- Crucial in conservation: They help assess the genetic health of populations, which is especially important in conservation biology.
Their diversity makes them perfect for a wide array of applications, from understanding evolutionary history to criminal investigations.
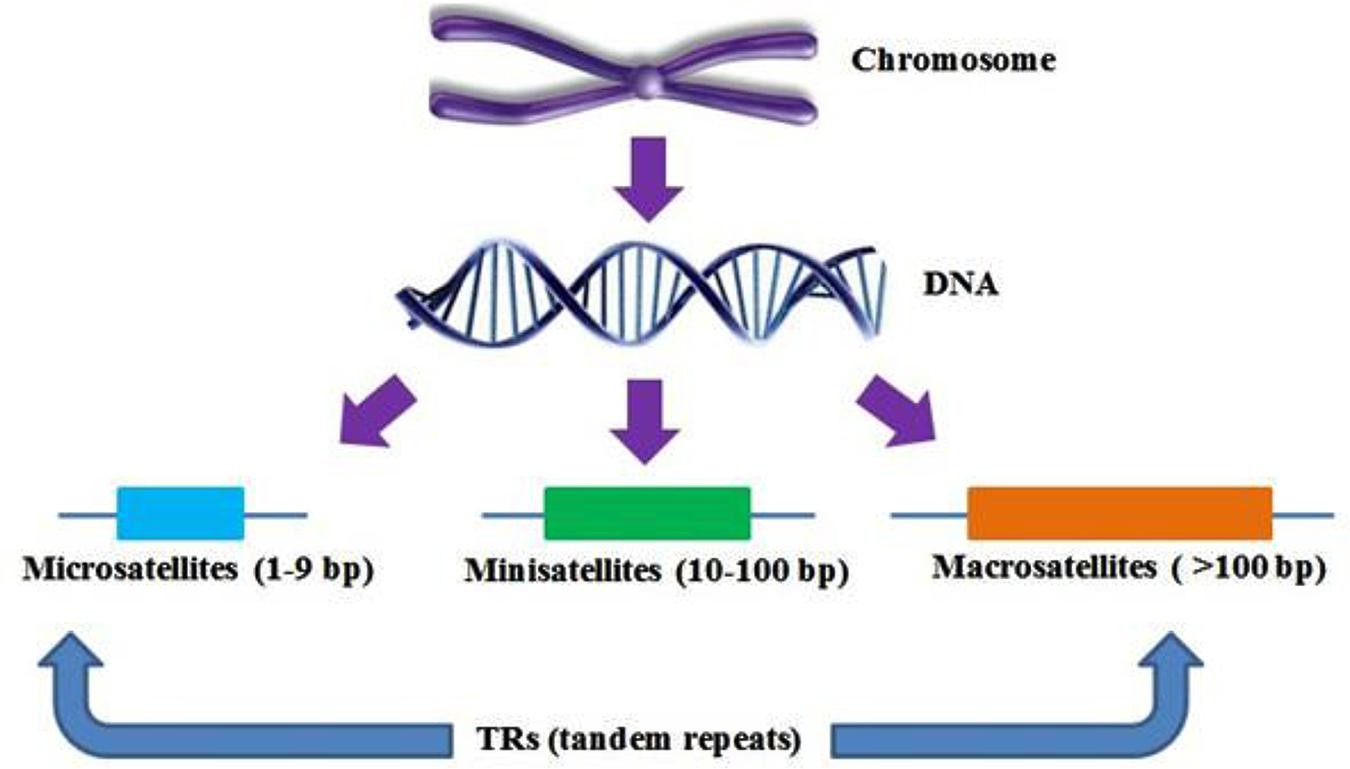 Diagram illustrating the different types of tandem repeats (TRs). (Abdullah F. Saeed et al,.2016)
Diagram illustrating the different types of tandem repeats (TRs). (Abdullah F. Saeed et al,.2016)
3. Restriction Fragment Length Polymorphisms (RFLPs)
Though a bit less common these days, RFLPs still have a seat at the table. These markers detect DNA variations by measuring differences in the size of DNA fragments after they're cut by restriction enzymes.
Why would you use RFLPs?
- Co-dominant: Like microsatellites, they can differentiate between heterozygotes and homozygotes.
- Genetic mapping: RFLPs were once the go-to tool for mapping genes in the early days of molecular genetics.
- Still handy in some applications: They're not as widely used as they once were, but in some genomic DNA sequencing studies, RFLPs still do the job.
RFLPs paved the way for modern genetic mapping and are still found in some genetic labs and studies today.
4. Amplified Fragment Length Polymorphisms (AFLPs)
AFLPs are another PCR-based marker, but they're a bit more complex. These markers work by amplifying various fragments from across the genome, with the goal of identifying genetic differences.
Why do researchers still use AFLPs?
- Dominant inheritance: This makes AFLPs a bit easier to interpret than co-dominant markers.
- High polymorphism: They provide plenty of genetic variation, which is helpful for studying population diversity.
- Moderate reproducibility: While AFLPs can provide solid data, sometimes the results aren't as consistent as you might want.
AFLPs remain useful in genetic diversity studies, particularly in population genetics, where understanding the genetic makeup of groups is key.
5. Random Amplified Polymorphic DNA (RAPD)
Last but not least, RAPD markers. These are also PCR-based and can be incredibly useful in genetic studies, but they have their quirks.
Here's what makes RAPDs tick:
Dominant inheritance: Similar to AFLPs, this makes RAPDs easier to interpret in some cases.
Polymorphism: They offer a good amount of genetic variation, though not always as much as other markers.
Low reproducibility: This can be a drawback because, if the experiment isn't controlled properly, you might not get consistent results every time.
Despite their low reproducibility, RAPDs have been valuable for rapid, preliminary studies of genetic diversity.
Summary of Comparison of Genetic Markers
| Marker Type |
Key Features |
Advantages |
Applications |
| Single Nucleotide Polymorphisms (SNPs) |
Smallest change in DNA sequence (one nucleotide difference) |
- Abundant and evenly distributed across the genome
- Easily automated for large-scale research
- Useful for understanding diseases and traits |
- Genome-wide association studies (GWAS)
- Population genetics
- Personalized genomics |
| Simple Sequence Repeats (SSRs) / Microsatellites |
Repeated DNA sequences of variable length |
- High polymorphism (genetic variation)
- Co-dominant inheritance (detects both copies)
- Powerful in breeding
- Useful for conservation studies |
- Forensic science
- Population genetics
- Conservation biology
- Marker-assisted selection (MAS) |
| Restriction Fragment Length Polymorphisms (RFLPs) |
DNA fragments cut by restriction enzymes |
- Co-dominant
- Useful in genetic mapping
- Historical importance in early genetic studies |
- Gene mapping
- Some genomic sequencing studies |
| Amplified Fragment Length Polymorphisms (AFLPs) |
PCR-based markers that amplify fragments from across the genome |
- High polymorphism
- Good for studying population diversity
- Easier interpretation due to dominant inheritance |
- Population genetics
- Genetic diversity studies |
| Random Amplified Polymorphic DNA (RAPD) |
PCR-based markers with no prior sequence knowledge needed |
- Dominant inheritance
- Offers genetic variation, though lower than other markers
- Quick and cost-effective |
- Preliminary genetic diversity studies
- Rapid assessment of genetic variation |
Genetic markers are like the toolbox for genetic research—each one with its own set of advantages and limitations. The choice of marker often depends on the specific goals of the study and the organism being investigated. While SNPs have become the go-to for many large-scale genomic studies, microsatellites still hold immense value in areas like forensics, conservation, and breeding programs. Other markers, like RFLPs, AFLPs, and RAPDs, may be less commonly used today but continue to offer valuable insights in certain contexts.
Ultimately, whether you're mapping a genome, investigating genetic diversity, or working on a genetic research project, understanding the strengths and weaknesses of these markers helps ensure you're using the right tool for the job.
For more in-depth information on microsatellites, including how they work in genetic research, CD Genomics offers a wide range of microsatellite-based services, from genotyping to instability analysis. Check out our services to learn more!
Microsatellite Markers vs. SNPs: A Comparative Analysis
Microsatellite Markers: A Deep Dive
Microsatellites, also known as Simple Sequence Repeats (SSRs), are short, repetitive sequences of DNA, typically 1-6 base pairs long, that are scattered throughout the genome. These repeats vary in length from one individual to another, making them perfect for studies that require high levels of polymorphism — that is, variations in the genetic code that differ between individuals.
Because of their highly variable nature, microsatellites are like the fingerprint of a species' DNA, providing a unique signature. These markers are invaluable in genetic diversity studies, paternity testing, and forensic applications. In fact, they're often used in forensic labs to link suspects to crimes or identify individuals through DNA profiling.
However, despite their usefulness, microsatellites do come with a set of challenges. Their high mutation rate, which makes them so versatile, can also make them more difficult to interpret in some contexts, as they may undergo rapid changes over generations. Still, when used appropriately, they offer unparalleled insight into genetic relationships and diversity.
SNPs: The Single Base Change
On the other hand, Single Nucleotide Polymorphisms (SNPs) are the most common type of genetic variation in humans. These markers involve a change in a single nucleotide—think of it as a single-letter typo in the vast book of the human genome. While SNPs might seem like small changes, their presence or absence in a genome can have significant consequences. A single nucleotide change can influence the function of a gene, affecting everything from susceptibility to diseases to physical traits.
The beauty of SNPs lies in their stability. Unlike microsatellites, which can be prone to slippage and mutation, SNPs are much more stable over time. This makes them perfect for population studies, genetic mapping, and pharmacogenomics (studying how genes affect drug response). With advances in genotyping technologies, SNPs have become the go-to tool for large-scale genetic research and personalized medicine.
Microsatellites vs. SNPs: The Battle of Markers
So, which one is better? The answer depends on the specific needs of your research. Let's break it down:
| Characteristic |
Microsatellites |
SNPs |
| Polymorphism |
High variability, great for diversity studies |
Less variation, better for population-wide studies |
| Mutation Rate |
High, more prone to changes |
Low, stable over generations |
| Length |
Typically 1-6 base pairs |
Single nucleotide change (1 base pair) |
| Cost & Technology |
Requires more complex detection methods |
Easier to detect, cost-effective for large-scale studies |
| Applications |
Forensics, paternity testing, biodiversity |
Disease studies, genetic mapping, drug response |
| Interpretation |
Can be tricky due to repeat slippage |
Easier to interpret due to stability |
When to Choose Microsatellites and SNPs
Microsatellites are ideal when:
- You need a highly polymorphic marker for individual identification or detailed genetic diversity analysis.
- You're working on forensic analysis or parentage testing.
- Your study involves species with complex genomes or requires fine-scale resolution of genetic variation.
SNPs are perfect when:
- You are interested in identifying genetic factors that contribute to disease or response to drugs.
- You're conducting large-scale genetic mapping studies.
- You want to perform population genetics studies with stable markers.
Real-World Statistics and Use Cases for Advantages of Using Microsatellite Markers
Even though SNPs are incredibly useful, microsatellites bring their own set of remarkable advantages, particularly in specific areas of research.
High Sensitivity and Precision
Microsatellites are known for their high polymorphism, which allows them to detect extremely fine-scale genetic differences between individuals. This makes them ideal for applications requiring precise genetic analysis, such as forensic identification and population genetics. For example, in wildlife conservation, microsatellites can help assess genetic diversity within a population, which is key to determining the genetic health of species. When studying tiger populations, microsatellite markers were used to identify genetically distinct groups, informing better conservation strategies.
Versatility Across Species
One of the most compelling features of microsatellites is their versatility. While SNPs are powerful, microsatellites can be used across a wide array of species, including non-model organisms—those that haven't been extensively studied or genetically mapped. This versatility is particularly advantageous in biodiversity monitoring and conservation genetics. For example, elephant conservation has benefitted from microsatellite analysis to assess genetic variation in fragmented populations, guiding conservation decisions based on genetic data, not just geography.
Conservation and Wildlife Monitoring
Microsatellites also excel in genetic fingerprinting, allowing researchers to track individuals, determine family relationships, and observe breeding patterns in wildlife populations. This is essential for conservation biology, as it helps inform decisions on species preservation and habitat protection. Through microsatellite analysis, researchers have gained insights into genetic diversity and population structure, helping protect species from the dangers of inbreeding and genetic bottlenecks.
At CD Genomics, we offer specialized services like microsatellite genotyping, microsatellite instability analysis, and microsatellite development. Learn more about our comprehensive offerings at Microsatellite Genotyping Service and Microsatellite Instability Analysis.
Disadvantages and Challenges of Microsatellite Markers
While microsatellites have plenty to offer, they are not without their challenges.
Complexity in Large Populations
One of the biggest drawbacks of microsatellites is their complexity in large-scale studies. Their repetitive structure means that scoring them accurately can be difficult. In large populations, where high variability exists in the number of repeat units, PCR amplification can face issues such as allelic drop-out or stutter bands, leading to false positives or miscalls. This makes microsatellites harder to analyze compared to SNPs, especially when working with large datasets.
Repeatability and Reliability
Due to the repetitive nature of microsatellites, their repeatability can be an issue. Mutational changes—such as allelic drop-out or incorrect allele interpretation—can occur during PCR amplification, leading to unreliable data. While SNPs are stable and generally resistant to these types of errors, microsatellites are more prone to such issues, requiring more rigorous quality control during analysis.
Applications of Microsatellite Markers in Research
Microsatellite markers play a crucial role in various fields of research:
- Genetic Diversity Studies: Used to assess genetic variation within and between populations.
- Marker-Assisted Selection: In agriculture, microsatellites are crucial for crop breeding programs to select desirable traits.
- Forensic Science: Microsatellites form the foundation of forensic DNA databases like CODIS, used in criminal investigations and paternity testing.
- Medical Research: Studies on genetic diseases like Huntington's disease and cancer rely on microsatellite markers to study mutation patterns and genetic predispositions.
For a deeper dive into the applications of microsatellites, check out our comprehensive guide to microsatellite markers here.
More applications, refers to What Are Microsatellite Markers Used For .
When to Choose Microsatellites Over Other Markers
Research Focus: Microsatellites vs. Other Markers
When diving into genetic research, especially in fields like forensics and biodiversity, microsatellite markers stand out as a highly valuable tool. Their high polymorphism allows for an exceptional level of sensitivity when detecting genetic differences at very fine scales, which is crucial for studies requiring high-resolution data.
For instance, in forensic DNA profiling, microsatellites have proven to be indispensable. Due to their high variability and repeat nature, they are excellent at distinguishing individuals. This unique ability makes them especially useful in parentage testing, criminal investigations, and species identification. A study by Jia et al. (2015) showcased how microsatellites were used for genetic fingerprinting in Chinese horses. The analysis helped identify genetic diversity and trace specific lineages, demonstrating the markers' effectiveness in fine-scale genetic studies (Jia, L., Zhang, Z., & Liu, Z., 2015).
Beyond forensics, microsatellites are also essential in genetic diversity studies, particularly when analyzing populations in both plants and animals. Sea turtles, for instance, are an iconic species for studying genetic structure through microsatellites. A study by Abreu et al. (2015) utilized microsatellite markers to examine the genetic differentiation of sea turtle populations, a task crucial for designing effective conservation strategies. The findings emphasized that microsatellites offer a deeper understanding of genetic variation at the population level, which other markers, like SNPs, may not capture with the same level of resolution (Abreu, L., et al., 2015).
When the research scale expands and the goal shifts towards large-scale genotyping or high-throughput sequencing, SNPs (Single Nucleotide Polymorphisms) are often the better choice. While microsatellites are perfect for detailed analysis of individual variation, SNPs provide a cost-effective and scalable approach for handling large volumes of genetic data across many samples.
SNPs are found throughout the genome, making them more accessible for genome-wide studies. Their stability over generations, combined with lower cost per marker, makes them ideal for studies like genetic mapping, population genetics, and pharmacogenomics. In a study by Wang et al. (2011), SNPs were successfully used for high-throughput analyses aimed at identifying disease-associated loci, proving that SNPs provide a robust solution for large genetic association studies (Wang, L., et al., 2011).
In addition, SNPs are widely employed in clinical research, as evidenced by a study by Yuan et al. (2016) that used SNP genotyping arrays to analyze genetic risk factors for diseases like breast cancer. The scalability of SNPs for high-throughput sequencing platforms, like next-generation sequencing (NGS), allows for the rapid analysis of large datasets, making them the go-to solution for genome-wide association studies (GWAS) and other large-scale clinical applications (Yuan, G.-C., et al., 2016).
While microsatellites provide exceptional detail in studies with specific objectives, SNPs excel in scenarios where scale, cost-effectiveness, and speed are key factors. CD Genomics, with its expertise in both microsatellite and SNP-based applications, offers tailored solutions to ensure that your study has the right tools for success. Whether you're investigating fine-scale genetic variation or conducting large genome-wide studies, CD Genomics can guide you toward the optimal choice for your research needs.
Ultimately, selecting the right genetic marker depends on the research goals. For detailed, fine-scale analyses like forensics or biodiversity studies, microsatellites are ideal. But for large-scale applications, such as genetic mapping, GWAS, or clinical research, SNPs offer scalability and efficiency. Understanding these differences helps researchers make informed decisions on the best approach for their specific objectives.
People Also Ask (PAA) Questions
What is the difference between microsatellite markers and SNP markers?
Microsatellites are repeat sequences that provide high variability, while SNPs are single-nucleotide variations and are more stable.
What are the advantages of microsatellite markers over other markers?
Microsatellites offer higher polymorphism, making them ideal for studying genetic diversity, forensic analysis, and population genetics.
Can microsatellite markers be used for forensic analysis?
Yes! STRs (a type of microsatellite) are fundamental in forensic DNA analysis, forming the basis for databases like CODIS.
Why are microsatellite markers important in population genetics?
Microsatellites help assess genetic diversity, structure, and evolutionary history within populations.
What are the disadvantages of using microsatellite markers?
Microsatellites can have high mutation rates, leading to reproducibility issues, and they are less scalable than SNPs.
Conclusion
In summary, microsatellite markers and other genetic markers like SNPs each have unique strengths and weaknesses. Microsatellites offer high polymorphism and are invaluable in forensic science, genetic diversity studies, and marker-assisted selection. However, for large-scale projects, SNPs may be more cost-effective and efficient.
If you are ready to enhance your research with the power of microsatellite markers, CD Genomics offers tailored services to meet your needs. From microsatellite genotyping to instability analysis and marker development, we are your trusted partner in genomic research.
References:
- Ellegren, H. (2004). Microsatellites: Simple sequences with complex evolution. Nature Reviews Genetics, 5(6), 435-445.
- Schneider, S., Roessli, D., & Excoffier, L. (2000). Arlequin: A software for population genetics data analysis. Computational Biology and Bioinformatics, 12(3), 712-732.
- Wang, J., & Guo, J. (2014). SNP genotyping: Technologies and applications in molecular genetics. Nature Biotechnology, 32(2), 125-132.
- Rafalski, A. (2002). Applications of single nucleotide polymorphisms in crop genetics. Current Opinion in Plant Biology, 5(2), 94-100.
- Jia, L., Zhang, Z., & Liu, Z. (2015). Microsatellite-based genetic fingerprinting for parentage testing and conservation of Chinese horses. Journal of Animal Science and Biotechnology, 6(1), 32.
- Abreu, L., dos Santos, A., & Andrade, R. (2015). Microsatellite markers reveal genetic structure and diversity of loggerhead sea turtle populations. Conservation Genetics, 16(3), 785-794.
- Wang, L., Song, H., & Liu, Q. (2011). Application of SNP markers in human genetic disease mapping. Human Genetics, 130(2), 185-195. · Yuan, G.-C., et al. (2016). Genetic variants associated with breast cancer risk identified through SNP-based analyses. Nature Genetics, 48(9), 1061-1072.
- Wang, J., et al. (2014). High-throughput sequencing and its applications in genetics and genomics. Trends in Biotechnology, 32(6), 356-365.


 Sample Submission Guidelines
Sample Submission Guidelines
 The upper DNA molecule differs from the lower DNA molecule at a single base-pair location (wikipedia)
The upper DNA molecule differs from the lower DNA molecule at a single base-pair location (wikipedia)  Diagram illustrating the different types of tandem repeats (TRs). (Abdullah F. Saeed et al,.2016)
Diagram illustrating the different types of tandem repeats (TRs). (Abdullah F. Saeed et al,.2016) 