The addition of methyl groups to specific DNA regions represents a fundamental epigenetic process that controls genetic expression patterns. Through this chemical modification, particularly when occurring at promoter sites, genes can be silenced without any changes to their underlying sequence. While promoter methylation typically suppresses gene activity, methylation within gene bodies often correlates with enhanced expression. This crucial regulatory mechanism orchestrates numerous biological processes, including cellular differentiation and immune system function.
Furthermore, it guides developmental trajectories across various tissues. When methylation patterns become disrupted, numerous pathological conditions may emerge, ranging from malignancies to disorders affecting neural function. To study these genome-wide methylation signatures effectively, researchers frequently employ specialized technologies like Infinium HumanMethylation935K platforms. These analytical tools have revolutionized our capacity to investigate regulatory networks and pathogenic processes by enabling comprehensive examination of methylation landscapes.
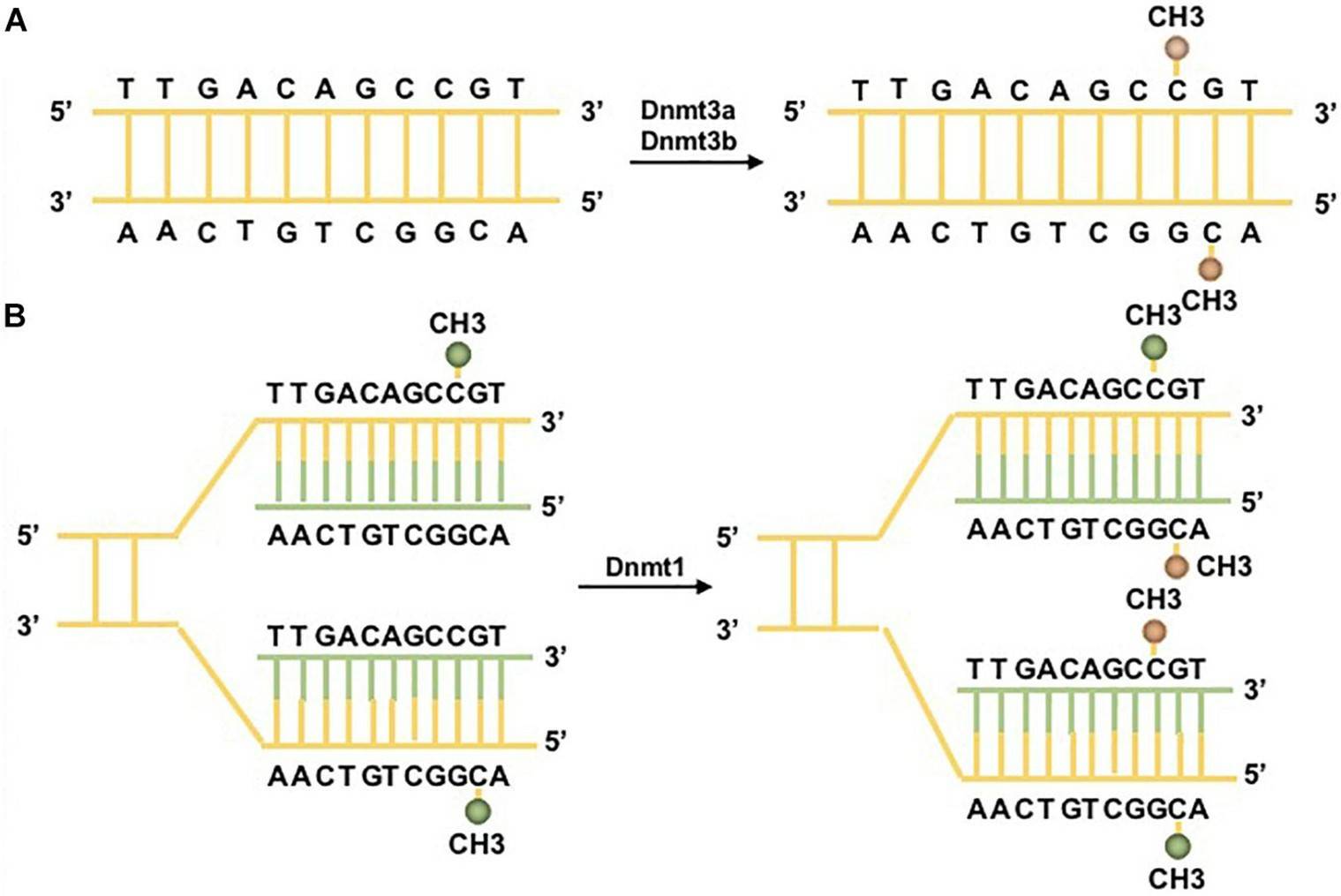 DNA methylation pathways. (Li, Jiaqi, et al. 2021)
DNA methylation pathways. (Li, Jiaqi, et al. 2021)
Early Methylation Arrays
The development of DNA methylation research was significantly shaped by initial microarray technologies, despite their technical constraints regarding coverage and precision. These pioneering arrays enabled researchers to examine DNA methylation configurations across selected CpG locations, which streamlined the concurrent evaluation of various specimens. Nevertheless, substantial drawbacks characterized these first-generation platforms. The primary constraints included restricted genomic scope and relatively imprecise quantification of methylation status, preventing thorough genome-wide assessment. Furthermore, the dependency on probe-based detection mechanisms introduced various technical challenges, including variable hybridization performance and experimental artifacts.
Subsequent technological progress brought forth enhanced platforms from Illumina, notably their Infinium series including the HumanMethylation450 and MethylationEPIC BeadChip systems. These advanced instruments represented a substantial leap forward in both analytical precision and genomic scope. The Infinium HumanMethylation450 technology achieved extensive coverage, encompassing the vast majority of both RefSeq genes and CpG islands. Building upon this foundation, the MethylationEPIC v2.0 system maintained similarly comprehensive coverage rates for these genomic elements.
Yet, contemporary high-density arrays continue to encounter certain methodological hurdles. Technical issues persist regarding probe functionality and binding efficiency, potentially affecting data quality. While foundational methylation array platforms were essential for advancing epigenetic understanding, their inherent limitations in scope and resolution restricted their broader scientific applications. Modern developments in sequencing methodology, particularly high-throughput approaches, are progressively addressing these historical constraints.
Advancements in Methylation Array Technology
Recent innovations in methylation detection platforms represent a significant breakthrough for studying genomic modifications, particularly in disease research and epigenetic investigations.
Understanding Dense Methylation Detection Platforms
Contemporary detection systems utilize sophisticated microarray technology to assess DNA methylation states, enabling researchers to examine numerous CpG locations simultaneously on individual chips. This capability generates detailed methylation landscapes across the genome. These modern platforms demonstrate significant improvements over previous generations, offering enhanced precision and expanded genomic coverage. Such technological capabilities support extensive epigenetic investigations while revealing complex relationships between genetic modifications and disease manifestations.
Platform Evolution and Enhanced Detection Capabilities
A notable milestone in methylation analysis came with Illumina's introduction of their advanced detection system, which incorporates refined Infinium I and II chemical processes. This platform examines in excess of 450,000 distinct CpG positions, with particular emphasis on crucial genomic elements including promoter regions, protein-coding sequences, and regulatory domains. Such widespread genomic sampling enables deeper insights into methylation distributions. The implementation of sophisticated reagents combined with refined analytical protocols has substantially improved measurement accuracy and reproducibility.
These state-of-the-art detection platforms have evolved into sophisticated instruments for investigating genomic methylation patterns, providing crucial information about epigenetic alterations and their role in pathological processes. The enhanced resolution and reliability of these systems make them invaluable tools for modern genomic research.
Service you may interested in
Infinium 450k Methylation Array
Infinium 450k Methylation Array: Design and Application
The design of the Infinium 450k Methylation Array incorporates two primary chemistries: Infinium I and Infinium II. Infinium I chemistry is responsible for detecting approximately one-third of the CpG sites, while Infinium II targets the remaining sites. This dual-chemistry approach not only enhances the coverage of CpG sites but also increases data stability and reproducibility. Furthermore, the array is well-suited for large-scale epigenetic studies, as exemplified by its use in The Cancer Genome Atlas (TCGA) project, where it facilitated the methylation analysis of over 75,000 samples.
 Infinium HumanMethylation450 BeadChip. Image source: Illumina.
Infinium HumanMethylation450 BeadChip. Image source: Illumina.
Impact of the Infinium 450k Methylation Array
The Infinium 450k Methylation Array has had a profound impact on methylation research. It enables large-scale epigenetic studies, allowing researchers to conduct extensive disease association analyses and broader epigenetic investigations. This technology has also propelled our understanding of complex diseases, providing insights into the pathogenesis of conditions such as cancer and neural tube defects. Additionally, the Infinium 450k array contributes significantly to early disease diagnosis, prognosis assessment, and the discovery of therapeutic targets, delivering critical data to these scientific endeavors.
Single CpG Site Resolution in Methylation Arrays
Contemporary methylation analysis techniques possess the capability to examine specific cytosine-guanine dinucleotides independently throughout genomic sequences. This granular analytical approach enables scientists to determine methylation percentages at discrete genomic positions, revealing sophisticated regulatory mechanisms and their physiological consequences.
Technological progress, especially regarding next-generation sequencing platforms like bisulfite-based genomic analysis methods (including both comprehensive and targeted approaches), has enabled detailed examination of DNA modification states at individual positions. These methodologies generate extensive sequence information with substantial depth, providing nuanced understanding of position-specific methylation patterns essential for deciphering regulatory mechanisms. Various analytical platforms, including specialized arrays and computational frameworks, have proven effective at accurately determining methylation levels at discrete genomic locations.
The capacity to examine individual modification sites holds particular importance because it enables detection of minor methylation variations that potentially influence transcriptional control, epigenetic modifications, and pathological processes. Within oncology investigations, modifications at specific positions often correlate directly with altered gene regulation, potentially triggering malignant transformation. Moreover, this precise analytical capability helps researchers elucidate how epigenetic modifications manifest across diverse cellular contexts and environmental settings, suggesting novel therapeutic strategies.
Position-specific methylation analysis represents more than improved measurement accuracy - it introduces revolutionary approaches for investigating DNA modifications and their biological roles. These methodological advances present significant opportunities for expanding knowledge in epigenetics and associated medical disciplines.
Infinium Methylation Arrays Beyond 450k
Since their inception, Infinium methylation arrays have undergone several iterations to enhance both resolution and coverage. The Infinium HumanMethylation450 BeadChip, commonly referred to as 450k, represents one of the earlier versions. It was capable of quantifying methylation states at approximately 485,577 CpG sites with single-base resolution. However, in response to increasing research demands, Illumina introduced chips with higher resolution, such as the Infinium HumanMethylationEPIC BeadChip (EPIC), extending coverage to approximately 850,000 CpG sites.
Improvements in Resolution and Coverage of Subsequent Chips
1. Infinium HumanMethylationEPIC BeadChip: Compared to the 450k, the EPIC chip offers higher resolution and broader coverage. It not only detects a greater number of CpG sites but also enhances data accuracy and reliability through the use of Infinium I and II probe designs.
2. Infinium 935K Chip: As one of the latest iterations, the 935K chip reportedly offers even higher resolution and coverage, while remaining compatible with various sample types and maintaining quality and reproducibility akin to the 450k.
 Infinium MethylationEPIC v2.0 (950k). Image source: Illumina.
Infinium MethylationEPIC v2.0 (950k). Image source: Illumina.
Technical Specifications and Research Applications
- 450k Chip: Covers approximately 485,577 CpG sites at single-base resolution, suited for large-scale genomic methylation analyses.
- EPIC Chip: Extends coverage to around 850,000 CpG sites, leveraging Infinium I and II technologies for enhanced detection accuracy and reliability.
- 935K Chip: Although details are sparse, it promises higher resolution and broader coverage, suitable for complex genomic analyses.
Research Applications
- 450k Chip: Widely used in fields like cancer and neurodegenerative disease research, offering comprehensive methylation profiling. Its lower probe density may not encompass all biologically relevant regions.
- EPIC Chip: With its superior resolution and coverage, EPIC is well-suited for high-precision methylation studies, including epigenetic research and exploration of complex disease mechanisms.
- 935K Chip: Expected to play a pivotal role in studies requiring high resolution and broad coverage, particularly in molecular epidemiology and complex disease research.
2024 Release: High-Throughput Methylation Analysis Tool
In 2024, Illumina launched the Infinium Methylation Screening Array-48 Kit, a high-throughput methylation analysis tool designed for large-scale population epigenomics studies.
 Infinium Methylation Screening Array (270k). Image source: Illumina.
Infinium Methylation Screening Array (270k). Image source: Illumina.
Technical Specifications
- Sample Capacity: Capable of processing up to 600,000 samples annually, making it ideal for extensive population studies.
- Probe Coverage: Covers approximately 270,000 unique methylation sites, focusing on CpG regions associated with human traits, disease phenotypes, environmental exposure, and aging.
- Throughput and Automation: Supports high throughput and automated liquid handling, significantly reducing per-sample cost and enhancing experimental efficiency.
- Data Quality: Demonstrates excellent reproducibility across more than 98% of samples.
Research Applications
- Population Epigenomics: Particularly suited for large-scale studies in common disease research, environmental epidemiology, and population genetics.
- Multi-Omics Research: Beyond methylation analysis, allows for non-CpG methylation and SNP analysis with high minor allele frequency.
- Clinical Applications: Applicable in cancer, aging, and molecular epidemiology research.
Comparison between Infinium 450k Chip and Infinium Methylation Screening Array
| Feature |
Infinium Methylation Screening Array-48 Kit |
Infinium HumanMethylation450 BeadChip (450k) |
| Coverage |
Approximately 270,000 methylation sites |
Approximately 485,000 CpG sites |
| Sample Capacity |
Capable of processing up to 600,000 samples per year |
Typically suited for small to medium-scale studies |
| Throughput & Automation |
High throughput with support for automated liquid handling; ideal for large-scale projects |
Low throughput with limited automation capabilities |
| Probe Quantity |
Around 270,000 unique methylation sites |
Approximately 485,577 CpG sites |
| Applicable Research |
Suitable for large-scale population studies, environmental epidemiology, and common disease research |
Large-scale research in cancer, neurodegenerative diseases, and epigenetics |
| Data Quality |
Good reproducibility across >98% of samples |
High quality, but constrained by probe density and throughput |
| Technical Platform |
Based on the EX Methylation platform, offering excellent automation and liquid handling |
Utilizes BeadArray technology, which is relatively complex |
| Cost Efficiency |
Lower sample cost, making it suitable for large-scale studies |
Higher cost, making it less feasible for ultra-large sample sizes |
| Technical Focus |
Focuses on specific CpG regions related to human traits, diseases, and aging |
Offers whole-genome coverage, suitable for comprehensive methylation analysis |
Services you may interested in
Want to know more about the details of DNA Methylation Arrays? Check out these articles:
Applications of High-Resolution Methylation Arrays
High-resolution methylation arrays find extensive applications across various domains, including cancer research, developmental biology, and environmental epigenetics. By providing precise analyses of DNA methylation patterns, these tools enable scientists to uncover critical insights into biological processes and disease mechanisms.
Cancer Research
In the realm of oncology, high-resolution methylation arrays are extensively utilized to identify genomic changes associated with tumorigenesis and cancer progression. For instance, using the Infinium HumanMethylation450 BeadChip platform, researchers can investigate methylation patterns linked to breast cancer and B-cell lymphoma, facilitating the identification of potential cancer biomarkers. Furthermore, these arrays are instrumental in studying DNA methylation alterations in lung cancer, revealing specific CpG island methylation patterns in early-stage squamous cell carcinoma of the lung. These studies are pivotal not only for understanding the molecular underpinnings of cancer but also for developing new strategies for early diagnosis and therapeutic interventions.
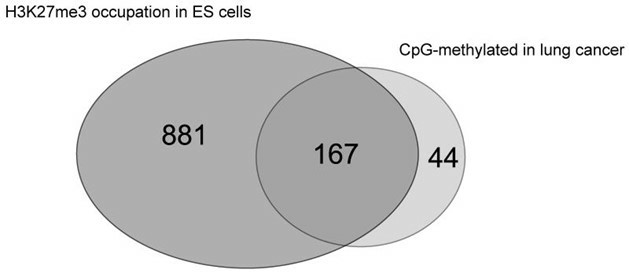 A large fraction of the methylated CpG islands in lung cancer is associated with the Polycomb complex in embryonic stem cells. (Pfeifer, et al., 2009.)
A large fraction of the methylated CpG islands in lung cancer is associated with the Polycomb complex in embryonic stem cells. (Pfeifer, et al., 2009.)
Developmental Biology
In developmental biology, high-resolution methylation arrays assist in elucidating the roles of gene expression regulation and epigenetic modifications throughout development. By analyzing genome-wide DNA methylation in mouse embryos, researchers can explore how epigenetic factors influence gene expression and cellular differentiation. This approach provides a vital tool for understanding the dynamic changes in gene regulatory networks during development.
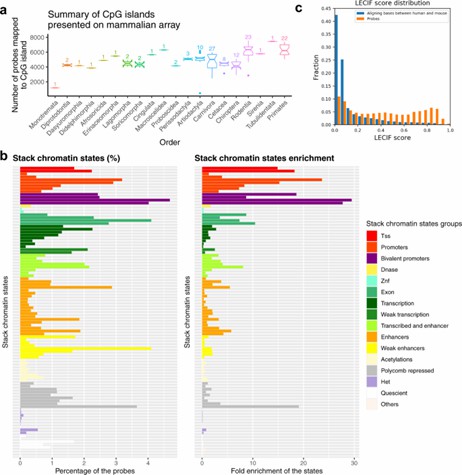 CpG island and chromatin state analysis of mammalian methylation probes. (Arneson, et al. 2022)
CpG island and chromatin state analysis of mammalian methylation probes. (Arneson, et al. 2022)
Environmental Epigenetics
Within the field of environmental epigenetics, high-resolution methylation arrays are employed to examine how environmental factors affect organisms through epigenetic mechanisms. For example, a study utilizing high-resolution methylation arrays analyzed DNA methylation patterns in the blood cells of asthma patients, uncovering associations between environmental exposures (such as smoking and air pollution) and the onset of asthma. This research sheds light on how environmental factors can impact health and disease by altering DNA methylation patterns.
In summary, high-resolution methylation arrays serve as critical tools for advancing our understanding of complex biological and environmental interactions, offering valuable insights for both fundamental research and potential clinical applications.
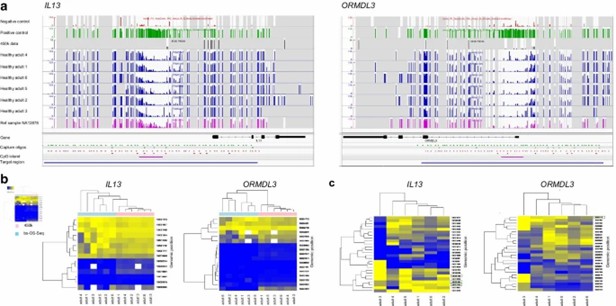 CpG methylation profiling of IL13 and ORMDL3 genes' islands and shores and comparison of bs-OS-Seq and 450k array in PBMC from healthy males. (Söderhäll, C., et al., 2021).
CpG methylation profiling of IL13 and ORMDL3 genes' islands and shores and comparison of bs-OS-Seq and 450k array in PBMC from healthy males. (Söderhäll, C., et al., 2021).
Want to know more about the details of DNA Methylation Arrays? Check out these articles:
Conclusion and Future Perspectives
Recent decades have witnessed remarkable advancements in DNA modification detection platforms, which have become essential instruments for studying epigenetic patterns and identifying disease markers. Nevertheless, researchers face various technical hurdles, including intricate computational requirements and inconsistent methodological approaches across laboratories. The irregular genomic distribution of modification sites combined with the complex nature of epigenetic information creates additional analytical difficulties, highlighting the necessity for unified experimental procedures and data processing guidelines to strengthen scientific reproducibility.
Looking ahead, technological innovations aim to enhance measurement precision down to individual base pairs, while simultaneously developing frameworks for integrating modification data with complementary molecular information from RNA expression and protein analyses. This integrated approach promises deeper understanding of biological systems. Novel applications continue emerging in clinical diagnostics and precision therapeutics, particularly regarding tumor-specific molecular signatures. Although technical and methodological challenges persist, these detection platforms maintain their position as fundamental tools for investigating health conditions and disease mechanisms.
References:
- Li, Jiaqi, et al. "Insights into the role of DNA methylation in immune cell development and autoimmune disease." Frontiers in Cell and Developmental Biology 9 (2021): 757318. https://doi.org/10.3389/fcell.2021.757318
- Wei, Rui, et al. "A Systematic Review of the Application of Machine Learning in CpG Island (CGI) Detection and Methylation Prediction." Current Bioinformatics 19.3 (2024): 235-249. https://doi.org/10.2174/1574893618666230508104341
- Alhassan, Daniel, Gayla R. Olbricht, and Akim Adekpedjou. "Differential methylation region detection via an array-adaptive normalized kernel-weighted model." Plos one 19.6 (2024): e0306036. https://doi.org/10.1371/journal.pone.0306036
- Pfeifer, Gerd P., and Tibor A. Rauch. "DNA methylation patterns in lung carcinomas." Seminars in cancer biology. Vol. 19. No. 3. Academic Press, 2009. doi: 10.1016/j.semcancer.2009.02.008
- Arneson, A., Haghani, A., Thompson, M.J. et al. A mammalian methylation array for profiling methylation levels at conserved sequences. Nat Commun 13, 783 (2022). https://doi.org/10.1038/s41467-022-28355-z
- Liu, Yang, et al. "High-spatial-resolution multi-omics sequencing via deterministic barcoding in tissue." Cell 183.6 (2020): 1665-1681. https://doi.org/10.1016/j.cell.2020.10.026
- Söderhäll, C., Reinius, L.E., Salmenperä, P. et al. High-resolution targeted bisulfite sequencing reveals blood cell type-specific DNA methylation patterns in IL13 and ORMDL3. Clin Epigenet 13, 106 (2021). https://doi.org/10.1186/s13148-021-01093-7
- Shen, J., LeFave, C., Sirosh, I. et al. Integrative epigenomic and genomic filtering for methylation markers in hepatocellular carcinomas. BMC Med Genomics 8, 28 (2015). https://doi.org/10.1186/s12920-015-0105-1


 Sample Submission Guidelines
Sample Submission Guidelines
 DNA methylation pathways. (Li, Jiaqi, et al. 2021)
DNA methylation pathways. (Li, Jiaqi, et al. 2021) Infinium HumanMethylation450 BeadChip. Image source: Illumina.
Infinium HumanMethylation450 BeadChip. Image source: Illumina. Infinium MethylationEPIC v2.0 (950k). Image source: Illumina.
Infinium MethylationEPIC v2.0 (950k). Image source: Illumina. Infinium Methylation Screening Array (270k). Image source: Illumina.
Infinium Methylation Screening Array (270k). Image source: Illumina. A large fraction of the methylated CpG islands in lung cancer is associated with the Polycomb complex in embryonic stem cells. (Pfeifer, et al., 2009.)
A large fraction of the methylated CpG islands in lung cancer is associated with the Polycomb complex in embryonic stem cells. (Pfeifer, et al., 2009.) CpG island and chromatin state analysis of mammalian methylation probes. (Arneson, et al. 2022)
CpG island and chromatin state analysis of mammalian methylation probes. (Arneson, et al. 2022) CpG methylation profiling of IL13 and ORMDL3 genes' islands and shores and comparison of bs-OS-Seq and 450k array in PBMC from healthy males. (Söderhäll, C., et al., 2021).
CpG methylation profiling of IL13 and ORMDL3 genes' islands and shores and comparison of bs-OS-Seq and 450k array in PBMC from healthy males. (Söderhäll, C., et al., 2021).