In the human genome, DNA methylation predominantly happens at cytosines of CpG sites, acting as an epigenetic modification that governs gene transcription and influences the maintenance of chromatin stability. This mechanism engages deeply in processes such as cellular development, embryonic differentiation, aging, and drug addiction. Moreover, DNA methylation might contribute to carcinogenesis through inhibition of tumor suppressor genes, playing a critical role in the incidences of autoimmune diseases, such as multiple sclerosis and diabetes. DNA methylation offers unique biological insights in cancer, neurological disorders, aging, and development, realms not approachable by other omics studies. Although methylation sequencing technology has been developed, methylation array technology maintains prominence within the public database due to its stability and convenience, providing data support for the majority of methylation research.
CD Genomics is dedicated to providing methylation array services for our clients, initiating services using the recent Methylation Screening Array (Illumina 270K). This platform furnishes researchers with a more cost-effective and reliable tool, propelling the profound progression of methylation research.
Introduction to Methylation Screening Array (MSA)
According to scholarly accounts, to achieve a precision comparable to that of methylation microarray, the depth of sequencing in whole-genome methylation must encompass at least 100×1 coverage. Since the advent of the Infinium Methylation Array, data exceeding 3 million cases has been accumulated globally, surpassing 20,000 related literatures and providing more than 225,000 public datasets for research inquiries. To prioritize the study of specific disease cohorts and broader health screenings, illumina has introduced a novel methylation microarray product named Methylation Screening Array (MSA).
The Infinium Methylation Screening Array covers approximately 270,000 methylation sites, known succinctly as MSA270K, with core applications in specific disease cohort research and extensive health screenings. The array primarily supports research focused on areas intrinsic to human traits, disease phenotype, environmental exposure, and aging. Ensuring high-level precision, this microarray has elevated the single array detection throughput by 48 samples, a six-fold increase from the Infinium MethylationEPIC v2.0, thereby achieving higher throughput and lower costs.
 Figure 1 Infinium Methylation Screening Array (image source illumina)
Figure 1 Infinium Methylation Screening Array (image source illumina)
Comparison of methylation assay 270K vs 935K
| Feature Comparison |
270k |
935k |
| Application |
- Common Disease Research |
- Cancer Research |
|
- Environmental Epidemiology |
|
|
- Population Genomics |
|
|
- Consumer Genomics |
|
|
- Rare Disease Research |
|
| Focus |
- Known associations of common diseases |
- Known associations of cancer |
| Key Focus Areas |
- Known associations of environmental exposures |
|
|
- Cell type-specific methylation |
|
|
- Intermediate methylation |
|
|
- Measurement of high MAF multi-functional SNPs |
|
|
- Single nucleotide polymorphisms |
|
| Coverage |
- Entire methylome coverage (>99%) |
- Comprehensive coverage of MGMT gene |
|
- RefSeq genes |
- Compatibility with published cancer classifiers |
|
- CNV detection |
- Compatibility with published rare disease classifiers |
|
- Comprehensive coverage of cancer-driving mutations |
|
| Chip Sites |
269,094 |
930,301 |
| Sample Throughput |
48 |
8 |
| Sample Input |
50ng |
250ng |
Trait-associated loci of Infinium Methylation Screening Array (MSA 270K)
MSA 270K incorporates an expansive number of disease-associated loci, stringently curated and validated. The chip encapsulates a comprehensive range of traits and disease manifestations spanning various fields including, but not limited to, cardiovascular, metabolic, mental health, autoimmunity, respiratory, reproductive, renal, aging, genetic, environmental exposure, and infectious domains.
| Trait Category |
Targeted Probes |
| Development/Aging |
102,533 |
| Environmental Exposure |
44,043 |
| Inflammation/Autoimmune Diseases |
41,894 |
| Ancestry |
31,843 |
| Gender |
23,806 |
| Infectious Diseases |
14,844 |
| Metabolic Diseases |
13,739 |
| Rare Genetic Diseases |
13,429 |
| Neurological/Neurodevelopmental Diseases |
8,874 |
| Physical Traits (Body Morphology) |
8,109 |
| Mental Disorders |
7,280 |
| Cardiovascular Diseases |
7,007 |
| Reproductive Biology/Health |
6,999 |
| Neurodegenerative Diseases |
4,733 |
| Lung/Respiratory Diseases |
1,748 |
| Renal Diseases |
982 |
Approximately 50% of known associated loci are derived from methylation array, these loci are deciphered through the analysis of published data, scientific literature, and the Infinium methylation array, aiding in the discovery of associations between CpG methylation and diverse traits or diseases.
Approximately 100K new CpG sites, primarily enriched in regulatory regions and cell-specific chromosomal states, are derived from whole-genome bisulfite sequencing (WGBS) data analyses at single-cell or bulk levels from published articles.
The array also promotes multi-omics research beyond CpG methylation. It incorporates an additional 2776 Cph sites and 3848 single nucleotide polymorphisms (SNPs with a high minor allele frequency (MAF)), selected from genomic databases.
Association enrichment comparison of traits in Infinium Methylation Screening Array (MSA 270K)
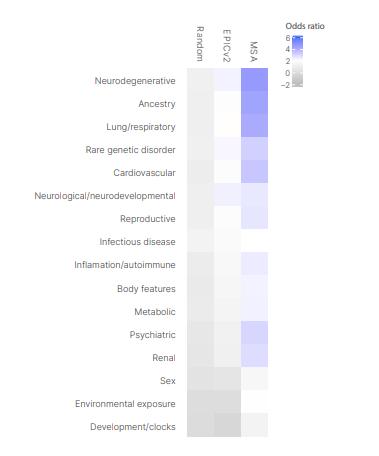 Figure 2 Markers on the Infinium Methylation Screening Array are highly enriched in known trait-associated CpGs across diverse trait types
Figure 2 Markers on the Infinium Methylation Screening Array are highly enriched in known trait-associated CpGs across diverse trait types
The MSA 270K Methylation array, leveraging cutting-edge Infinium array chemistry, implements multiple bead-based retests for each CpG site. Each bead carries thousands of probes to ensure the accuracy and reliability of the detection results. After stringent internal testing, the reproducibility of the MSA 270K exceeds 98%, while its overlapping probes with the Infinium Methylation EPIC v2.0 (935K) exhibit reproducibility of over 96%, demonstrating exceptional performance and stability.
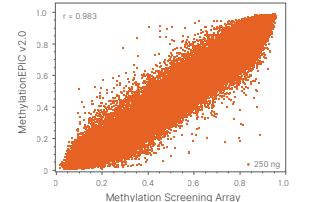
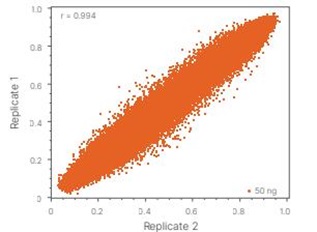 Figure 3 Infinium Methylation Screening Array performance
Figure 3 Infinium Methylation Screening Array performance
Advantage of Methylation Screening Array
Focused Coverage: The array targets 270K unique methylation sites associated with common human traits, disease phenotypes, exposures, aging, and more. This focused coverage ensures comprehensive exploration of relevant epigenetic variations.
Scalability: With the capacity to process up to 600K samples per year on a single iScan™ System using the 48-sample BeadChip, the array enables large-scale methylation projects with ease.
Reliable Data: The array provides reliable methylation data with over 98% sample-to-sample reproducibility, ensuring consistency and accuracy in research findings.
Expert Content Selection: Content selection for the array is based on expert curation, incorporating powerful trait associations from published studies, functional genomics sequencing, and genomic databases. This ensures that the array covers known and predicted epigenetic associations comprehensively.
Multiomic Capabilities: Beyond CpG methylation, the array also targets non-CpG methylation sites and includes interrogation of single nucleotide polymorphisms (SNPs) with high minor allele frequencies. This multiomic approach provides unique insights into disease mechanisms across diverse populations.
Novel Content from WGBS: The array includes content selected from whole-genome bisulfite sequencing (WGBS) data sets, targeting loci associated with various genomic features, including cell type, gene expression, chromatin accessibility, and more. This novel content enhances the array's utility in exploring epigenetic landscapes.
Improved Scalability with Methylation Workflow: Powered by the Infinium EX Methylation platform, the array offers improved scalability and cost-effectiveness for high-volume methylation studies. The advanced workflow reduces processing costs and turnaround time, making it suitable for large population-level projects.
Simple QC and Data Analysis: Illumina provides software tools for quality control analysis of array data, ensuring reliable results.
Workflow of Methylation Screening Array Analysis
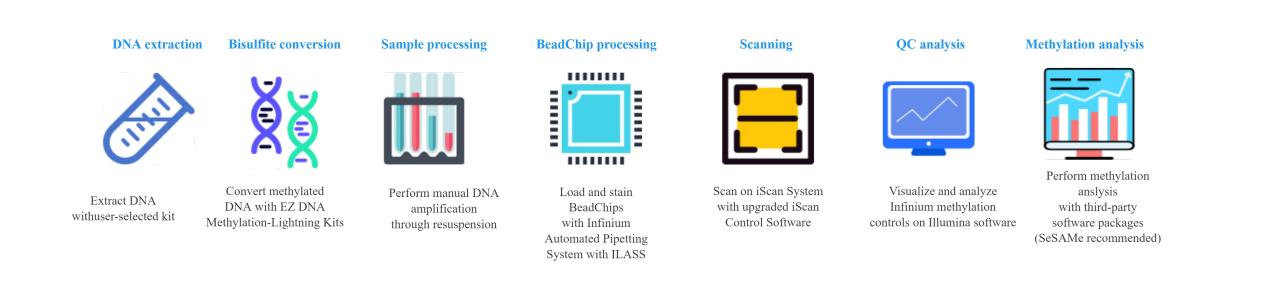
Bioinfomatics analysis
| Analysis |
Features |
| Standard |
Standard files provided: |
|
- Sample annotation |
|
- Variable annotation |
|
- Scanner output (IDAT files) |
| Differential Methylation Analysis |
Detection of variations in methylation levels among different sample groups. |
|
Files provided: |
|
- Comprehensive report summarizing findings and visualization plots |
|
- Data file containing identified differentially methylated CpGs, with breakdown by region |
|
(e.g., CpG islands, shelves, shores, open sea) |
|
- File containing differential methylated regions (DMRs), highlighting significant methylation changes |
| Gene Ontology Terms Analysis |
Explore enrichment of gene sets through analysis of Gene Ontology terms. |
|
Identification of biological processes potentially influenced by differential methylation. |
| Pathway Analysis |
Unveil biochemical pathways enriched with genes affected by differential methylation. |
|
Identify potential biological mechanisms underlying observed methylation variations. |
Delivery
Raw Data
Probe Report
Peaks Report
Summary Report
Other Available illumina DNA Methylation Array
| Microarray |
Species |
Coverage |
Platform |
| Infinium Human Methylation 850k |
Human |
Over 850,000 methylation sites per sample at single-nucleotide resolution. |
Illumina |
| Infinium Human Methylation 935K |
Human |
Detects over 935,000 CpG sites, covering CpG islands, promoters, coding regions, and enhancers comprehensively. |
Illumina |
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.


 Sample Submission Guidelines
Sample Submission Guidelines
 Figure 1 Infinium Methylation Screening Array (image source illumina)
Figure 1 Infinium Methylation Screening Array (image source illumina) Figure 2 Markers on the Infinium Methylation Screening Array are highly enriched in known trait-associated CpGs across diverse trait types
Figure 2 Markers on the Infinium Methylation Screening Array are highly enriched in known trait-associated CpGs across diverse trait types
 Figure 3 Infinium Methylation Screening Array performance
Figure 3 Infinium Methylation Screening Array performance
