Introduction of DNA Methylation and Illumina Arrays
The covalent attachment of methyl groups to DNA represents a fundamental epigenetic alteration, occurring predominantly at cytosine residues within CpG dinucleotides. Orchestrated by methyltransferase enzymes, this biochemical modification serves multiple biological functions, from modulating transcriptional activity to ensuring proper embryogenesis and maintaining chromosomal architecture. Although traditionally viewed as a repressive mark, methylation occasionally enhances gene activity in specific contexts. The preservation of genome integrity and suppression of potentially harmful mutations depend significantly on proper DNA methylation patterns, whose disruption frequently correlates with various pathological states, particularly neoplastic transformation.
Since their emergence in the early 2000s, methylation detection platforms developed by Illumina have revolutionized epigenetic research capabilities. These innovative tools enable comprehensive analysis of methylation signatures across the genome by identifying stable epigenetic modifications at CpG-rich regions, particularly accessible in blood samples. The evolution of these technologies, from the HumanMethylation450 platform to the enhanced coverage provided by the EPIC BeadChip system, has established new standards in methylation profiling.
These sophisticated analytical platforms serve dual purposes in both fundamental scientific investigation and medical applications. Researchers have successfully employed these arrays to elucidate how environmental factors, including nutrition and tobacco use, influence epigenetic modifications. The technology continues advancing through development of specialized sequencing chips, expanding our capacity to decode the complexities of epigenetic regulation. Their applications extend into clinical diagnostics, offering novel approaches for disease screening and risk evaluation.
Service you may interested in
Want to know more about the details of DNA Methylation Arrays? Check out these articles:
Evolution of Illumina Methylation Arrays
The landscape of epigenetic investigation has been transformed by sophisticated methylation detection platforms, which enable researchers to conduct extensive analyses of epigenetic modifications throughout the genome. Illumina's continuous technological refinements have substantially enhanced the capabilities, precision, and economic efficiency of methylation detection systems over the past ten years.
Chronological Development of Detection Platforms
The field witnessed a pivotal advancement in 2010 when researchers gained access to the Infinium 450K system, which facilitated examination of 450,000 distinct CpG locations. A significant expansion occurred in 2015 with the release of the MethylationEPIC v1.0, extending analytical reach to encompass 850,000 CpG sites and strengthening coverage of gene regulatory elements.
The year 2020 brought innovation through specialized arrays designed for model organism research, particularly in murine studies. Subsequently, 2021 saw the introduction of parallel processing capabilities through the Custom BeadChip 24-Kit, facilitating concurrent analysis of multiple samples. Further technological progression led to the 2022 release of MethylationEPIC v2.0, which expanded coverage to 935,000 CpG sites while improving analytical precision. Recent developments include the 2024 launch of screening arrays optimized for population-scale investigations.
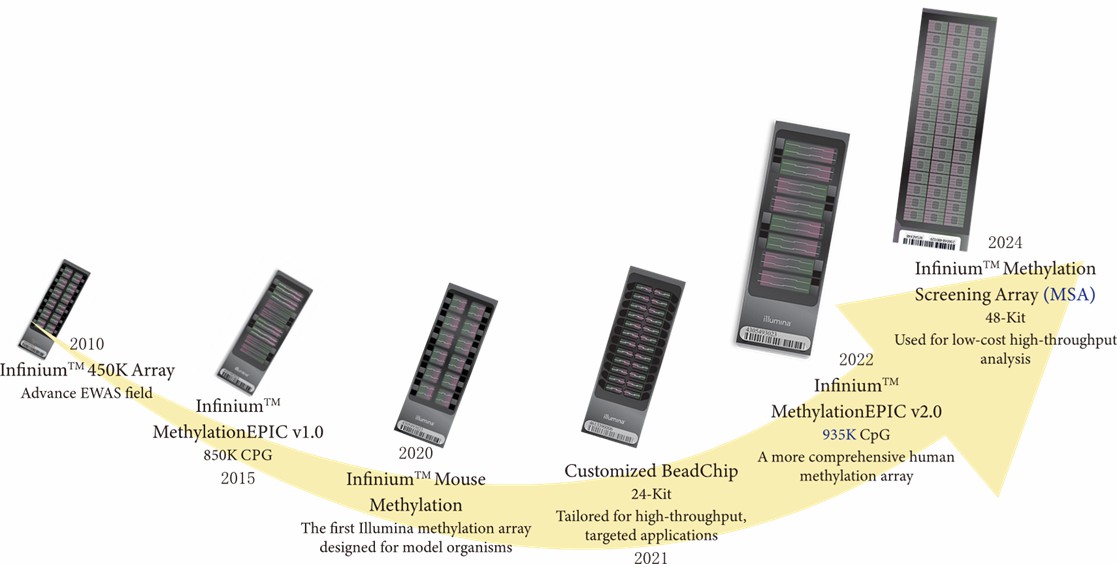 The development history of Illumina methylation arrays. Chip image source: Illumina.
The development history of Illumina methylation arrays. Chip image source: Illumina.
Infinium 450k Methylation Array
The Infinium HumanMethylation450 BeadChip, commonly referred to as the 450k chip, was Illumina's first widely adopted methylation array, released in 2011. It covers approximately 485,000 CpG sites, primarily located in key regulatory regions such as gene promoters, enhancers, and CpG islands, making it an essential tool in epigenetic research. Despite its relatively limited coverage, its cost-effectiveness, high throughput capabilities, and reliable data quality made it the preferred instrument for early epigenome-wide association studies (EWAS).
Infinium MethylationEPIC v1.0 850k CpG
Subsequently, Illumina launched the Infinium MethylationEPIC v1.0 array, also known as the 850k chip. This advancement was marked by the addition of approximately 363,753 new CpG sites, bringing total coverage to about 853,377 CpG sites. The 850k chip thus offers more comprehensive coverage of significant genomic regions such as enhancers, CTCF binding sites, and CNV detection areas. Offering higher resolution and broader genomic coverage compared to the 450k chip, the 850k chip is well-suited for more complex epigenetic studies.
Infinium MethylationEPIC v2.0 935k CpG
The latest iteration, the Infinium MethylationEPIC v2.0 array, or the 935k chip, further extends its coverage with the inclusion of over 86,000 additional CpG sites, reaching approximately 935,000 CpG sites in total. These newly added probes encompass enhancer-targeted regions, super-enhancers, CTCF binding sites, and CpG islands associated with cancer driver mutations. The 935k chip not only enhances data quality but also improves the detection capabilities for specific biomarkers, offering a wide range of applications in oncology, complex disease, and developmental research.
Infinium Methylation Screening Array 48-Kit (270K)
In response to the demand for large-scale screening, Illumina has also introduced the Infinium Methylation Screening Array 48-Kit, also known as the 270k chip. This array provides a high-throughput methylation analysis solution at a lower cost, suitable for large-scale epidemiological studies and preliminary screening efforts. Although its coverage is less extensive than previous generations, it still delivers sufficient information for initial epigenetic research inquiries.
Overall, these advancements in Illumina's methylation array technologies continue to significantly contribute to the evolution and expansion of epigenomic research.
Service you may intersted in
High-Throughput Methylation Arrays Analysis
Revolutionary developments in methylation detection platforms have redefined possibilities within epigenetic investigations, dramatically enhancing research capabilities and experimental scope. Such technological progress has catalyzed significant breakthroughs in genome-wide methylation studies, offering researchers powerful tools to decode epigenetic modifications associated with diverse phenotypes and pathological conditions.
Advantages of High-Throughput Technologies
Optimized Processing and Scale
Advanced methylation detection systems, particularly those developed by Illumina, facilitate simultaneous examination of numerous biological specimens, revolutionizing the temporal and financial aspects of epigenetic research. These platforms surpass traditional methodologies by enabling parallel processing of vast sample quantities at reduced operational costs. This capability proves essential for investigating complex biological systems where extensive sampling is crucial for meaningful insights.
Implementation in Comprehensive Epigenetic Investigations
Contemporary methylation analysis platforms serve as foundational tools for extensive epigenetic studies, encompassing diverse research domains from population-level genomic variation to environmental influences on gene regulation. These sophisticated analytical methods allow scientists to conduct detailed examinations of methylation signatures, yielding valuable insights into disease predisposition, wellness parameters, and the subtle interplay between genetic factors and environmental conditions. Such technological capabilities have proven instrumental in advancing our understanding of epigenetic mechanisms.
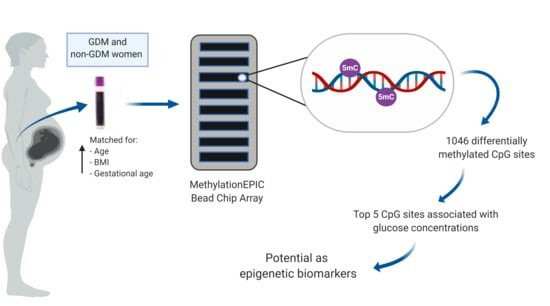 Altered Genome-Wide DNA Methylation in Peripheral Blood of South African Women with Gestational Diabetes Mellitus. (Dias, Stephanie, et al. 2019)
Altered Genome-Wide DNA Methylation in Peripheral Blood of South African Women with Gestational Diabetes Mellitus. (Dias, Stephanie, et al. 2019)
Impact on EWAS Development
Advancing Epigenetic Biomarker Discovery
Modern methylation detection platforms enable systematic examination of DNA modifications across entire genomes, revolutionizing our ability to connect epigenetic signatures with clinical manifestations. These analytical systems help scientists uncover numerous methylation changes that correlate with disease states, potentially serving as diagnostic indicators or therapeutic targets. Such capabilities significantly advance personalized treatment approaches.
Understanding Disease Complexity
Contemporary methylation analysis techniques have transformed investigations into challenging medical conditions, yielding particularly valuable results in cancer research, heart disease studies, and neurological disorders. Scientists now compare methylation profiles between patient cohorts and reference populations to decode epigenetic aspects of pathological processes. These methodologies prove especially valuable when examining conditions like metabolic disorders and obesity, where multiple genetic elements interact with environmental variables. Through comprehensive methylation studies, researchers gain crucial understanding about the molecular mechanisms underlying these intricate biological processes.
The implementation of these advanced analytical approaches continues generating novel insights into disease mechanisms, facilitating more precise diagnostic strategies and targeted therapeutic interventions. This technological progress has fundamentally altered our approach to studying complex medical conditions, offering unprecedented views into their epigenetic foundations.
For more details, please refer to the following articles:
Future Directions and Innovations in Methylation Microarray Technology
Emerging Trends in Methylation Microarray Technology
As technological advancements persist, methylation microarray technology is poised to evolve toward higher resolution, broader coverage, and increased sensitivity. Future iterations of methylation arrays are anticipated to enhance CpG site coverage, particularly in low-frequency or challenging regions such as GC-rich areas and repetitive sequences. Additionally, with the continuous improvement in sequencing technologies, methylation arrays may increasingly integrate with next-generation sequencing (NGS) platforms. This integration could further enhance data accuracy and depth, facilitating cost-effective, high-throughput methylation analysis at single-cell or individual levels.
Furthermore, future methylation arrays are likely to become more intelligent, incorporating sophisticated automation tools and machine learning algorithms. These advancements will assist researchers in swiftly pinpointing disease-associated methylation markers within vast datasets, potentially playing a critical role in the early diagnosis and prevention of conditions such as cancer, neurodegenerative diseases, and complex traits.
Integration with Other Genomic Technologies
The increasing availability of multi-omics data underscores the importance of integrating methylation microarray technology with other genomic approaches in future studies. By synergizing methylation data with genomics, transcriptomics, and proteomics datasets, researchers can gain comprehensive insights into gene expression regulation networks and their roles in diverse biological processes. For example, by combining methylation and gene expression data, scientists can better understand the impact of methylation on gene transcription, thereby elucidating the intricate relationships between gene regulation and disease pathogenesis.
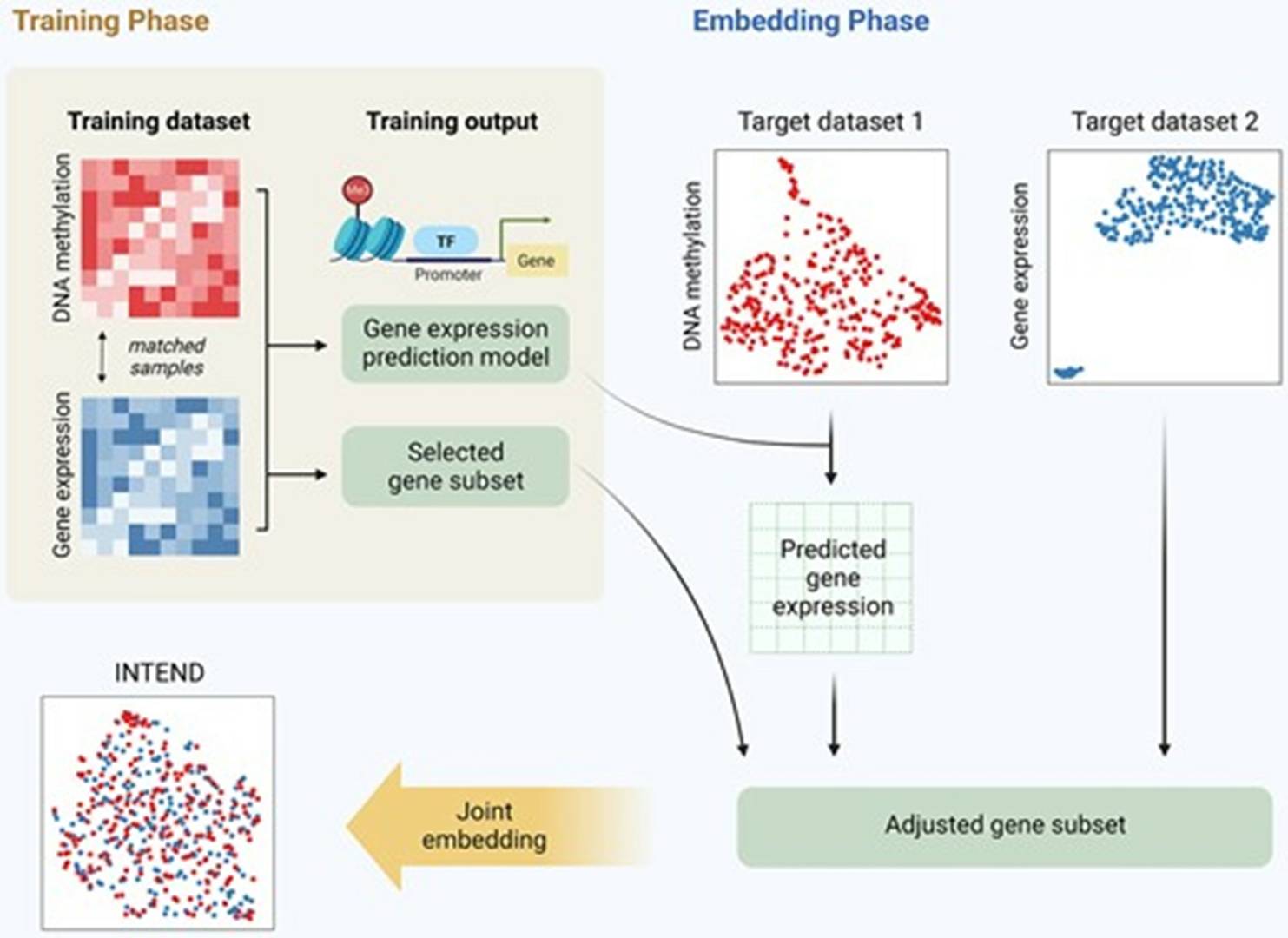 Integration of gene expression and DNA methylation data across different experiments. (Itai, et al., 2023)
Integration of gene expression and DNA methylation data across different experiments. (Itai, et al., 2023)
Service you may intersted in
Want to know more about the details of DNA Methylation Arrays? Check out these articles:
Conclusion
This article has reviewed the evolution of Illumina's methylation microarray technology, tracing its development from the earliest Infinium 450K Array to the latest Infinium Methylation Screening Array 48-Kit. Each generation of these arrays has contributed significantly to advancements in epigenetic research. Throughout this progression from the 450K to the 850K and then to the 935K chips, both genomic coverage and data quality have been continuously enhanced. This progression has allowed researchers to conduct more refined analyses of a greater number of CpG sites, thereby advancing the field of EWAS.
These technological breakthroughs have not only led to the identification of disease-associated epigenetic markers but have also played a crucial role in large-scale epidemiological studies.
With continued advancements, customizable arrays like the 24-Kit and the cost-effective Methylation Screening Array 48-Kit have provided viable solutions for large-scale screening and targeted research efforts, thereby expanding the scope of methylation array applications. Both fundamental research and clinical studies benefit from the robust support offered by Illumina's methylation array technology in the application of epigenetics.
Looking forward, methylation array technology is set to develop towards higher resolution, broader coverage, and reduced costs. The integration of single-cell methylation analysis with NGS technologies will usher in a new era of precision and personalized data analysis in methylation research. Furthermore, the convergence of methylation arrays with other omics technologies, including genomics, transcriptomics, and proteomics, will provide more comprehensive insights into the complex mechanisms underlying diseases, thereby advancing precision medicine and personalized treatment approaches.
As technological innovations persist, future methylation research is poised to play an increasingly significant role in the early diagnosis of diseases and the discovery of biomarkers, while also providing deeper insights into areas such as gene-environment interactions. Overall, the future of methylation array technology holds exciting potential and is expected to continue being a cornerstone in epigenetic research, driving progress in the life sciences and medicine.
References:
-
Umer, Muhammad, and Zdenko Herceg. "Deciphering the epigenetic code: an overview of DNA methylation analysis methods." Antioxidants & redox signaling 18.15 (2013): 1972-1986. https://doi.org/10.1089/ars.2012.4923
- Dias, Stephanie, et al. "Altered genome-wide DNA methylation in peripheral blood of South African women with gestational diabetes mellitus." International Journal of Molecular Sciences 20.23 (2019): 5828. https://doi.org/10.3390/ijms20235828
- Itai, Yonatan, Nimrod Rappoport, and Ron Shamir. "Integration of gene expression and DNA methylation data across different experiments." Nucleic Acids Research 51.15 (2023): 7762-7776. https://doi.org/10.1093/nar/gkad566


 Sample Submission Guidelines
Sample Submission Guidelines The development history of Illumina methylation arrays. Chip image source: Illumina.
The development history of Illumina methylation arrays. Chip image source: Illumina. Altered Genome-Wide DNA Methylation in Peripheral Blood of South African Women with Gestational Diabetes Mellitus. (Dias, Stephanie, et al. 2019)
Altered Genome-Wide DNA Methylation in Peripheral Blood of South African Women with Gestational Diabetes Mellitus. (Dias, Stephanie, et al. 2019) Integration of gene expression and DNA methylation data across different experiments. (Itai, et al., 2023)
Integration of gene expression and DNA methylation data across different experiments. (Itai, et al., 2023)