The modification of DNA through methylation represents a vital epigenetic mechanism that significantly shapes gene expression by adding methyl groups onto specific nucleotides - primarily cytosine and adenine. As a key regulatory process, methylation orchestrates multiple biological functions: it controls gene activity, facilitates the inactivation of one X chromosome, helps maintain genomic integrity, and guides cell differentiation pathways. When methylation patterns become disrupted, various pathological conditions can emerge, ranging from malignant transformations to impaired neural function and abnormal development. Understanding these methylation dynamics proves crucial for elucidating genetic regulation mechanisms and developing innovative therapeutic strategies.
This review aims to provide scientists with detailed guidance for identifying optimal methylation analysis platforms suited to their experimental objectives. The complexity inherent in methylation analysis techniques makes the appropriate selection of analytical methods critical for generating high-quality, dependable results. Our discussion encompasses an evaluation of different methylation detection platforms and their specific uses, providing investigators with crucial knowledge to select methodologies that align with their unique research requirements.
Understanding Methylation Arrays
Overview of Methylation Detection Technology
A genome-wide methylation detection platform employs sophisticated microarray technology to quantify DNA methylation patterns across numerous genomic locations. These platforms incorporate vast arrays of sequence-specific probes targeting CpG locations, enabling simultaneous evaluation of methylation states at multiple genomic positions. Among the available technologies, Illumina's platforms stand out, with progressive iterations including their initial 27K design, the expanded 450K version, and the current EPIC system, each offering increasingly comprehensive genomic coverage for methylation analysis.
Technical Principles and Methodology
The fundamental mechanism relies on sequence-specific binding interactions between sample DNA and immobilized oligonucleotide sequences. These detecting elements are anchored to microscopic silica spheres positioned within array wells. Following DNA-probe interaction, fluorescent-tagged nucleotides complement the target sequences, generating measurable signals. Advanced laser detection systems quantify the emitted fluorescence, which correlates directly with site-specific methylation levels. Researchers typically express these measurements as beta values, derived through the calculation β=M/(M+U), where M indicates methylated signal intensity and U represents unmethylated signal strength. This high-capacity analytical approach enables efficient methylation profiling across numerous specimens, supporting diverse applications in oncology, genetic research, and epigenetic investigations.
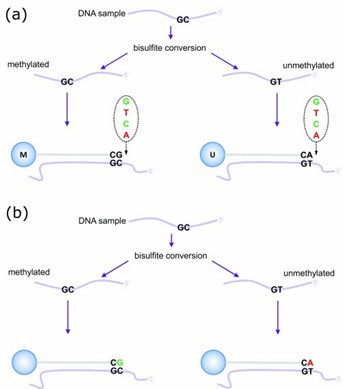 Illumina Infinium HumanMethylation450 assay. (Maksimovic, et al., 2017)
Illumina Infinium HumanMethylation450 assay. (Maksimovic, et al., 2017)
Service you may interested in
Want to know more about the details of DNA Methylation Arrays? Check out these articles:
Types of DNA Methylation Arrays
1. Illumina Methylation Arrays
Features and Advantages of Illumina Arrays
Illumina methylation arrays are highly regarded for their high-throughput capabilities, enabling the simultaneous analysis of numerous samples with single-nucleotide resolution. Central to this capability is the Infinium Methylation Assay, which allows precise quantification of DNA methylation at targeted CpG sites. This precision is critical for unraveling gene regulatory mechanisms and exploring epigenetic modifications linked to diseases such as cancer. These arrays offer comprehensive genomic coverage, including key regions like enhancers and CpG islands, making them versatile tools for epigenetics and molecular biology research.
Different Models Available
The Illumina array portfolio includes several models designed to meet diverse research requirements:
- Infinium MethylationEPIC BeadChip: Targets approximately 930,000 unique methylation sites, providing extensive coverage for cancer research and genetic studies.
- Infinium HumanMethylation450K BeadChip: Focuses on 485,000 probes, widely used for general methylation profiling.
- Infinium Custom Methylation Kit: Allows the design of custom arrays to target specific regions of interest.
- Infinium Mouse Methylation BeadChip: Tailored for epigenetic research in mouse models.
Coverage and Resolution of Illumina Arrays
Illumina arrays are distinguished by their broad coverage and high resolution. The Infinium MethylationEPIC v2.0 array includes over 930,000 probes, many of which target enhancer regions and other biologically significant genomic areas. This model improves upon the previous 450K array by incorporating around 350,000 additional probes, thereby enhancing the capacity to capture diverse methylation patterns across various tissues. The high resolution facilitates detailed insights into the methylome, essential for biomarker identification and understanding disease mechanisms.
2. Affymetrix Methylation Arrays
Characteristics of Affymetrix Arrays
Affymetrix methylation arrays are engineered to offer comprehensive DNA methylation analysis across the genome. They employ a distinctive probe design for the simultaneous interrogation of multiple methylation sites, noted for their high sensitivity and specificity in distinguishing methylated from unmethylated DNA.
Comparison of Coverage and Resolution with Illumina Arrays
Typically, Affymetrix arrays feature lower probe density compared to Illumina's high-throughput offerings. For example, while Illumina's Infinium MethylationEPIC addresses around 930,000 sites, Affymetrix arrays generally cover fewer sites but may deliver distinct data types or analytical methodologies. This variance can impact platform selection based on specific research objectives or sample types.
Unique Features or Benefits
A notable strength of Affymetrix arrays lies in their capability to integrate with other genomic data types, promoting multi-omics approaches in research. Furthermore, Affymetrix provides robust analytical tools that enhance data interpretation and integration with existing datasets. Although they may not match the probe abundance of Illumina arrays, these unique features offer significant advantages in contexts that require integration with diverse genomic analyses.
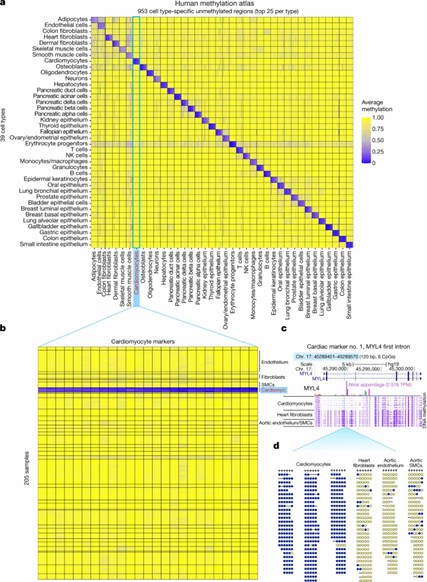 A human methylation atlas of 205 samples across 39 cell type groups. (Loyfer, et al., 2023)
A human methylation atlas of 205 samples across 39 cell type groups. (Loyfer, et al., 2023)
Factors to Consider in Methylation Array Selection
1. Research Goals
Influence of Specific Research Questions on Array Choice
Selecting an appropriate methylation array is fundamentally guided by the research questions at hand. For research focused on investigating associations with prevalent diseases, targeted arrays such as the Infinium Methylation Screening Array may be the most suitable due to its concentrated coverage of 270,000 methylation sites associated with known traits and diseases. On the other hand, comprehensive genome-wide studies may benefit from the Infinium MethylationEPIC BeadChip, offering extensive coverage of over 930,000 sites.
Examples of Research Goals and Suitable Arrays
- Population Health Studies: The Infinium Methylation Screening Array is optimized for large-scale epigenome-wide association studies (EWAS) targeting common diseases.
- Cancer Research: The Infinium MethylationEPIC array provides the needed comprehensive coverage for in-depth cancer-related methylation profiling.
- Environmental Exposure Studies: Arrays focusing on trait associations, such as the Screening Array, are effective for investigating the effects of environmental factors on DNA methylation.
2. Sample Type and Quality
Impact of Sample Type on Array Selection
The type of biological sample used is critical in the selection of a methylation array. Blood samples, which often yield high-quality DNA, are generally suitable for most arrays. In contrast, formalin-fixed paraffin-embedded (FFPE) tissue samples, which may contain degraded DNA, necessitate arrays specifically designed to handle this condition. The Infinium MethylationEPIC array supports a range of sample types including blood and FFPE tissues, while the Screening Array is tailored for low-input samples.
Considerations for Samples with Varying DNA Quality
For samples with varied DNA quality, researchers should select arrays capable of low-input or degraded DNA analysis. The Infinium Methylation Screening Array, requiring only 50 ng of DNA, is advantageous for studies facing limitations in sample quantity or quality.
3. Cost
Comparison of Costs of Different Methylation Arrays
Cost is a pivotal consideration in methylation array selection. The Infinium Methylation Screening Array is optimized for cost-effectiveness in high-throughput studies, enabling the processing of up to 16,128 samples weekly at a lower cost compared to the Infinium MethylationEPIC array.
Trade-offs Between Cost and Features
Cost-effective options may entail compromises in coverage and resolution. For instance, while the Screening Array offers specific application-focused content, it lacks the extensive probe coverage necessary for detailed genomic investigations present in the EPIC array.
Cost-saving Options or Bulk Purchase Deals
To optimize budgets, researchers should investigate bulk purchase deals or discounts for high-volume orders offered by suppliers. Such options can significantly reduce per-sample costs in large-scale studies.
4. Data Analysis and Support
Importance of Data Analysis Capabilities
Sophisticated data analysis capabilities are essential when selecting a methylation array. The complexity of methylation data requires advanced analysis tools to accurately interpret results and derive meaningful insights.
Comparison of Data Analysis Tools and Support Available for Different Arrays
Illumina provides a comprehensive suite of software tools for data analysis from their arrays, including quality control features and visualization tools via the GenomeStudio Methylation Module. In comparison, Affymetrix offers user-friendly analysis resources with distinct functionalities and support nuances.
User-friendly Resources or Tutorials
Both Illumina and Affymetrix provide online resources and tutorials that benefit researchers. For instance, Illumina recommends Bioconductor packages such as SeSAMe, which aid in the downstream analysis of methylation data. These resources enhance usability and facilitate navigation of complex data sets.
5. Summary
Choosing the appropriate methylation array involves a careful evaluation of research objectives, sample type and quality, cost factors, and the support available for data analysis. By aligning these considerations with specific research requirements, scientists can optimize study designs and enhance their findings in the field of epigenetic research.
| Factor |
Consideration |
Recommended Array(s) |
| Research Goals |
Disease/Trait Studies |
Infinium Methylation Screening Array (270,000 sites) |
|
Comprehensive Studies |
Infinium MethylationEPIC BeadChip (930,000 sites) |
|
Population Health Studies |
Infinium Methylation Screening Array |
|
Cancer Research |
Infinium MethylationEPIC BeadChip |
| Sample Type and Quality |
High-quality DNA (e.g., blood) |
All arrays, including MethylationEPIC |
|
Degraded DNA (e.g., FFPE) |
MethylationEPIC, Screening Array |
|
Low DNA Input |
Infinium Methylation Screening Array (50 ng) |
| Cost |
High-Throughput, Cost-effective |
Infinium Methylation Screening Array |
|
Detailed Analysis |
Infinium MethylationEPIC BeadChip |
| Data Analysis and Support |
Advanced Analysis Tools |
Illumina GenomeStudio, Bioconductor packages |
|
User-Friendly Resources |
Illumina and Affymetrix tutorials and tools |
Case Studies
Between 2013 and 2023, numerous case studies have highlighted the successful application of Illumina and Affymetrix methylation array technologies. Below are several notable studies and their findings:
1. Applications of the Illumina Platform:
- In 2014, a study utilized the Illumina 450K methylation array to analyze brain tissue from individuals with autism compared to controls. The study uncovered disruptions in various biological pathways and found a notable inverse correlation between methylation levels and gene expression of immune response-related genes in the autism group.
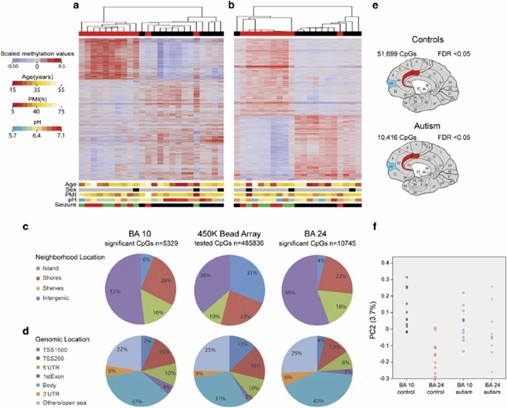 DNA methylation changes in autistic cerebral cortex regions. (Nardone, et al., 2014)
DNA methylation changes in autistic cerebral cortex regions. (Nardone, et al., 2014)
- Another investigation employed the Illumina Infinium HumanMethylation450 array to assess DNA methylation in 48 adult males and 35 adult females, demonstrating the platform's reliability in detecting DNA methylation.
- In 2020, researchers evaluated the data integration processes of Illumina 450K and EPIC platforms, offering effective data processing methods for epidemiological research.
2. Applications of the Affymetrix Platform:
- In breast cancer research, a study successfully transitioned clinically relevant gene expression features from Affymetrix microarrays to Illumina RNA sequencing, exemplifying cross-platform application.
- Another study compared results from Affymetrix and Illumina microarray platforms, finding that while some systematic biases existed, appropriate normalization methods could attenuate these differences.
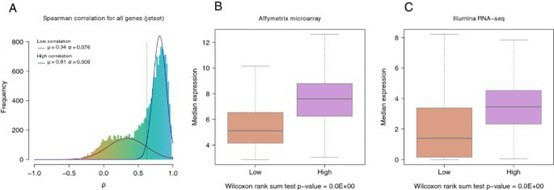 Gene expression correlation between Affymetrix microarray and Illumina RNA-Seq platforms. (Fumagalli, et al., 2014)
Gene expression correlation between Affymetrix microarray and Illumina RNA-Seq platforms. (Fumagalli, et al., 2014)
3. Cross-Platform Comparisons and Integration:
- A comparative analysis explored the relationship between gene methylation data and mRNA expression values across NimbleGen, Affymetrix, and Illumina platforms. Despite differences in data distribution across platforms, the overall trends remained consistent.
- In a cross-platform study involving multiple platforms, combining data from Affymetrix and Illumina microarrays enhanced statistical power and facilitated robust re-analyses.
These studies demonstrate the extensive utility of Illumina and Affymetrix methylation array technologies across various research fields. With careful processing and analytical approaches, these platforms can effectively illuminate biological mechanisms and disease-related pathways. These successful case studies provide valuable guidance and insights for future research endeavors.
Want to know more about the details of DNA Methylation Arrays? Check out these articles:
Conclusion
In this review, we deliver an in-depth analysis regarding the critical considerations that guide methylation platform selection. Our examination delves into the fundamental principles and applications of genomic methylation detection technologies, with particular emphasis on their capacity to measure epigenetic modifications throughout the genome. Through detailed evaluation of various platforms from manufacturers including Illumina and Affymetrix, we explore distinctive capabilities and relative strengths, enabling investigators to identify optimal solutions for their experimental goals.
Selecting appropriate methylation analysis technology profoundly impacts research outcomes and data reliability. Success depends on careful evaluation of multiple variables: experimental design requirements, biological specimen integrity, financial constraints, and computational analysis capabilities must align with the chosen platform. Thorough consideration of these elements facilitates generation of meaningful results that contribute substantively to epigenetics research advancement.
References:
- Tran, Q.T., Breuer, A., Lin, T. et al. Comparison of DNA methylation based classification models for precision diagnostics of central nervous system tumors. npj Precis. Onc. 8, 218 (2024). https://doi.org/10.1038/s41698-024-00718-3
- Leti, F., Llaci, L., Malenica, I., DiStefano, J.K. (2018). Methods for CpG Methylation Array Profiling Via Bisulfite Conversion. In: DiStefano, J. (eds) Disease Gene Identification. Methods in Molecular Biology, vol 1706. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-7471-9_13
- Jaunmuktane, Z., Capper, D., Jones, D.T.W. et al. Methylation array profiling of adult brain tumours: diagnostic outcomes in a large, single centre. acta neuropathol commun 7, 24 (2019). https://doi.org/10.1186/s40478-019-0668-8
- Maksimovic, Jovana, Belinda Phipson, and Alicia Oshlack. "A cross-package Bioconductor workflow for analysing methylation array data." F1000Research 5 (2017): 1281. doi: 10.12688/f1000research.8839.3
- Loyfer, N., Magenheim, J., Peretz, A. et al. A DNA methylation atlas of normal human cell types. Nature 613, 355–364 (2023). https://doi.org/10.1038/s41586-022-05580-6
- Nardone, S., Sharan Sams, D., Reuveni, E. et al. DNA methylation analysis of the autistic brain reveals multiple dysregulated biological pathways. Transl Psychiatry 4, e433 (2014). https://doi.org/10.1038/tp.2014.70
- Vanderlinden, L.A., Johnson, R.K., Carry, P.M. et al. An effective processing pipeline for harmonizing DNA methylation data from Illumina's 450K and EPIC platforms for epidemiological studies. BMC Res Notes 14, 352 (2021). https://doi.org/10.1186/s13104-021-05741-2
- Fumagalli, D., Blanchet-Cohen, A., Brown, D. et al. Transfer of clinically relevant gene expression signatures in breast cancer: from Affymetrix microarray to Illumina RNA-Sequencing technology. BMC Genomics 15, 1008 (2014). https://doi.org/10.1186/1471-2164-15-1008


 Sample Submission Guidelines
Sample Submission Guidelines Illumina Infinium HumanMethylation450 assay. (Maksimovic, et al., 2017)
Illumina Infinium HumanMethylation450 assay. (Maksimovic, et al., 2017) A human methylation atlas of 205 samples across 39 cell type groups. (Loyfer, et al., 2023)
A human methylation atlas of 205 samples across 39 cell type groups. (Loyfer, et al., 2023) DNA methylation changes in autistic cerebral cortex regions. (Nardone, et al., 2014)
DNA methylation changes in autistic cerebral cortex regions. (Nardone, et al., 2014) Gene expression correlation between Affymetrix microarray and Illumina RNA-Seq platforms. (Fumagalli, et al., 2014)
Gene expression correlation between Affymetrix microarray and Illumina RNA-Seq platforms. (Fumagalli, et al., 2014)