In genetic research, precise genetic markers are essential for understanding species traits, genetic diversity, and evolutionary mechanisms. Microsatellite markers, due to their high variability, widespread distribution across genomes, and efficiency in genetic studies, have become indispensable in genomics. Whether in crop improvement, species conservation, or forensic science, microsatellite markers demonstrate immense potential. However, as research demands grow, traditional methods of microsatellite marker development are struggling to meet the high efficiency and high-throughput requirements of modern scientific endeavors.
 Distribution of microsatellite markers across the genome. (Elizabeth Bryda et al,.2008)
Distribution of microsatellite markers across the genome. (Elizabeth Bryda et al,.2008)
To address these challenges, innovations in microsatellite marker development have become critical. By incorporating advanced Next-Generation Sequencing (NGS) technologies and sophisticated bioinformatics tools, the discovery and application of microsatellite markers have become faster, more accurate, and cost-effective.
This article will explore the significance of microsatellite marker development, starting with a comparison between traditional methods and the latest NGS technologies. We will then delve into the bioinformatics tools that are revolutionizing microsatellite marker discovery. Finally, we will look ahead to how these innovations are driving forward microsatellite development and pushing the boundaries of genetic research.
Importance of Microsatellites in Genetic Research
Microsatellites are pivotal in various fields of genetic research, including:
- Genetic mapping for identifying genes associated with diseases.
- Population genetics, where they help measure genetic variation within and between populations.
- Forensic science, where they are widely used in DNA profiling.
- Biodiversity conservation, where they help monitor genetic diversity in endangered species.
Microsatellite Marker Development: An Overview
What is a Microsatellite Marker?
Microsatellite marker development refers to the process of identifying and creating specific genetic markers based on microsatellites. Microsatellites are short, repetitive sequences of DNA, typically consisting of 1 to 6 base pairs, that are scattered throughout an organism's genome. These repetitive regions are highly variable between individuals, which makes them excellent candidates for genetic markers.
Steps Involved in Microsatellite Marker Development
The development of microsatellite markers typically follows several stages:
1. Microsatellite Discovery: Identifying microsatellite loci from genome sequences using sequencing technologies such as Next-Generation Sequencing (NGS). You can learn more about NGS and how it helps in microsatellite detection here.
2. Primer Design: Specific primers are designed for the microsatellite loci to amplify the DNA region. Tools like MISA or SSRLocator can be helpful for primer design.
3. Validation: Testing the primers on different samples to check for cross-species amplification and polymorphism.
4. Optimization: Refining PCR conditions to ensure robust and reproducible amplification across different samples.
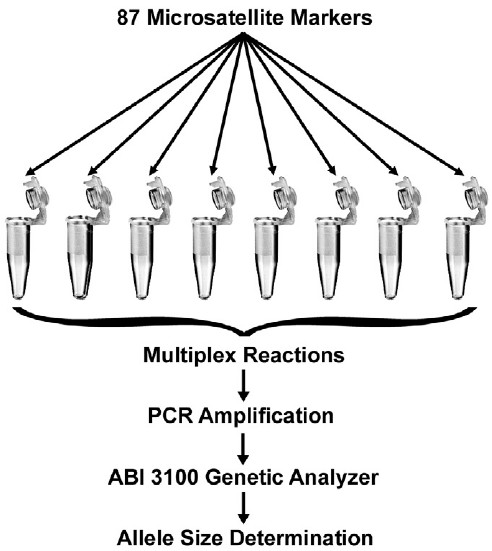
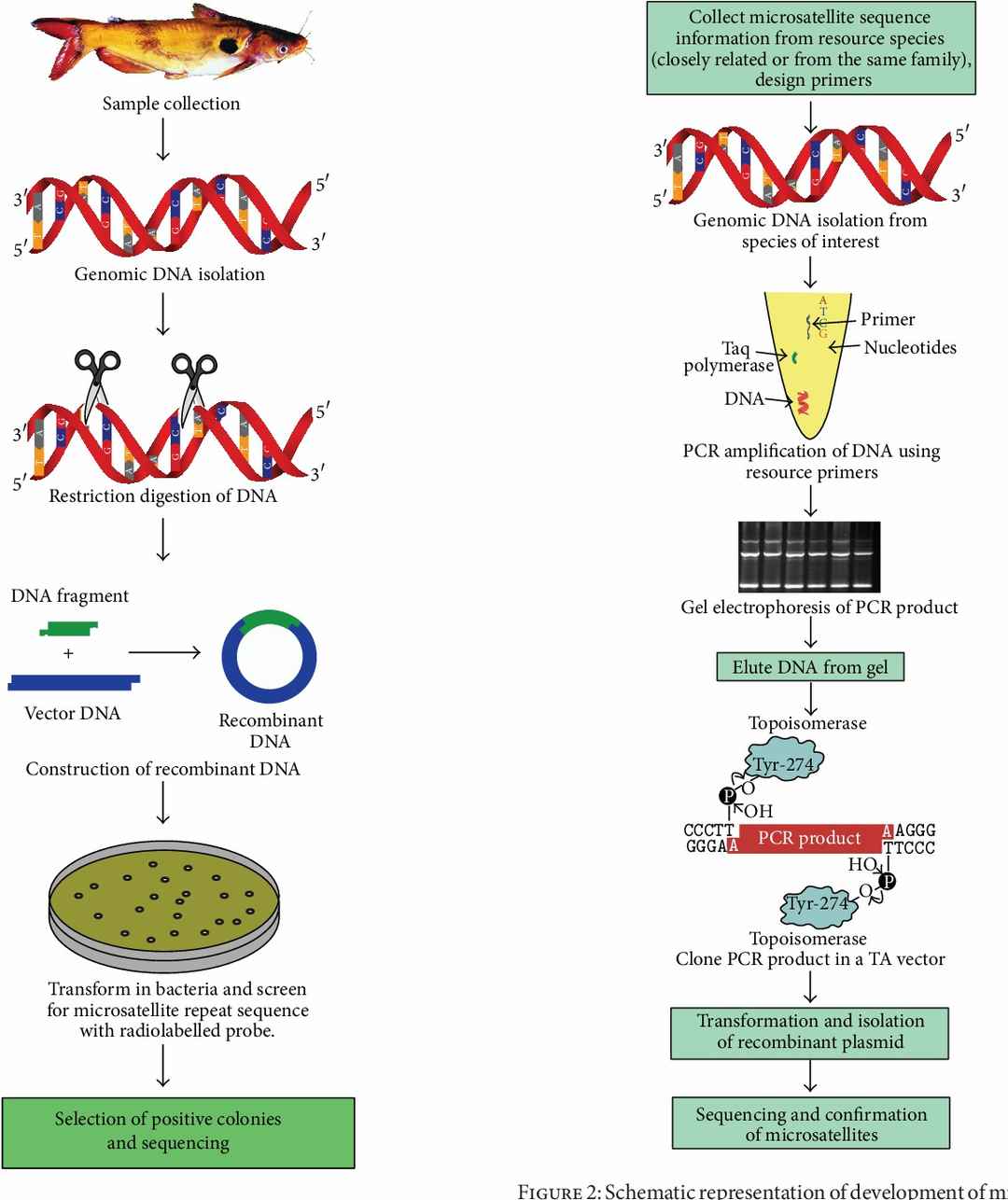 Schematic representation of development of microsatellite markers by cross-species amplification. (P. M. Abdul-Muneer et al,.2014)
Schematic representation of development of microsatellite markers by cross-species amplification. (P. M. Abdul-Muneer et al,.2014)
Applications of Microsatellite Markers
Use in Genetic Mapping
Microsatellite markers are crucial for genetic mapping in both plants and animals. They enable the identification of genes linked to important traits such as disease resistance, yield in crops, or specific characteristics in livestock. For instance, microsatellite genotyping is often used in plant breeding to select for traits like drought tolerance and pest resistance.
Learn more about our Microsatellite Genotyping Service for in-depth genetic mapping.
Microsatellites are extensively used in population genetics to evaluate genetic diversity, population structure, and evolutionary patterns. By analyzing microsatellite variability, researchers can trace gene flow, genetic drift, and other evolutionary processes within natural populations.
Microsatellites in Forensic Science and Biodiversity Studies
Forensic science relies heavily on microsatellites due to their ability to distinguish individuals with high accuracy. DNA profiling through microsatellites is one of the most reliable methods for forensic identification.
In biodiversity studies, microsatellites help assess the genetic health of endangered species by identifying genetic bottlenecks and estimating population sizes.
Methodologies for Microsatellite Marker Development
Traditional Methods vs. Next-Generation Sequencing (NGS)
Traditional methods for developing microsatellite markers, notably Sanger sequencing, have long been the standard approach. However, while Sanger sequencing provides highly accurate results, it is burdened with several limitations that make it less efficient in large-scale applications:
Time-Consuming: Traditional sequencing is labor-intensive, requiring extensive time for each locus to be sequenced individually. For example, the process of designing and sequencing markers for each microsatellite locus across multiple samples can take several weeks to months, depending on the sample size and complexity of the genome (Zhao et al., 2012). This can be particularly challenging in genomic studies requiring high-density marker maps.
Expensive: Sequencing each locus one by one is expensive, especially when dealing with large numbers of markers or species with complex genomes. Costs can rise significantly due to the necessity of using specialized equipment and reagents for each sequencing step. Studies have indicated that the cost of using Sanger sequencing for marker development in species like Pinus sylvestris can reach upwards of $5,000 per species (Valsecchi et al., 2014).
In contrast, Next-Generation Sequencing (NGS) has dramatically transformed the landscape of microsatellite marker development, offering several clear advantages:
Higher Throughput: NGS platforms such as Illumina and Ion Torrent allow the simultaneous sequencing of thousands of DNA fragments, including multiple microsatellite loci, in parallel. This increases throughput exponentially compared to traditional sequencing, enabling rapid discovery of markers. A study on rice (Oryza sativa) demonstrated that NGS could sequence an entire genome, identify over 10,000 microsatellite loci, and reduce the time for marker discovery to just a few weeks (Mather et al., 2018).
Cost-Effective: The ability to sequence large genomic regions at a reduced cost per base pair is a significant advantage of NGS. According to a study by Elshire et al. (2011), the cost per marker using NGS platforms is lower than traditional methods, especially for large-scale projects. This has made it possible for researchers to develop hundreds or even thousands of markers at a fraction of the cost, enabling large-scale genomic studies in crop breeding and conservation.
Accuracy: NGS technology provides deeper sequencing coverage and better accuracy when detecting polymorphisms, particularly in complex genomes. Unlike Sanger sequencing, which may miss rare alleles or fail to detect polymorphisms across populations, NGS offers greater sensitivity and precision. For example, a study by Wang et al. (2018) found that NGS technologies consistently detected more microsatellite markers in complex plant genomes compared to Sanger sequencing, improving marker reliability.
An example of the efficiency of NGS is seen in agricultural genomics. Researchers have successfully used NGS to identify hundreds of drought resistance markers in maize (Zea mays), which could be integrated into breeding programs to improve crop resilience (Zhang et al., 2017). This rapid marker discovery reduces costs associated with field trials and accelerates the breeding process.
For a more detailed discussion of microsatellite instability in genomic analysis, check out our Microsatellite Instability Analysis page.
Real-World Statistics and Use Cases for
Case Study of Microsatellites in Population Bioinformatics Approaches in Microsatellite Discovery
The integration of bioinformatics tools has made microsatellite marker development faster, more efficient, and more accurate. Rather than manually searching for microsatellites within large genomic datasets, bioinformatics tools enable automated detection and primer design, significantly improving the speed of discovery.
MISA (Microsatellite Identification Tool): MISA is widely used for detecting microsatellites in genomic data by scanning for repeat sequences. It automates the process of identifying candidate loci for marker development. Studies have shown that MISA can rapidly identify high-quality microsatellites from a variety of species, including plants like Brachypodium distachyon (Huang et al., 2012), reducing the time spent on manual searches.
QDD (Quantitative Data Descriptor): QDD is another key bioinformatics tool that aids in the analysis of microsatellite loci. It provides the ability to quantify polymorphism data and select the most informative loci for population studies. QDD is particularly useful for studies in population genetics and evolutionary biology, where variability is key (Ralph et al., 2015). For example, QDD has been used to identify microsatellite markers in Arabidopsis thaliana, contributing to a better understanding of genetic diversity in wild populations (Steinkellner et al., 2015).
MSTRX: This tool is designed to assist in the primer design process, an essential step for PCR amplification. MSTRX helps design specific primers that target microsatellite loci, ensuring high-quality amplification in experimental setups. Its utility has been demonstrated in studies such as Yu et al. (2017), where MSTRX was used to optimize primer design for poplar trees, aiding in the development of markers associated with disease resistance traits.
Furthermore, tools like RepeatMasker and SSRLocator further streamline the analysis by eliminating repetitive genomic sequences that are not relevant for microsatellite marker development (Liu et al., 2013). These tools ensure that only true microsatellites are selected for downstream analysis, improving marker specificity.
A case study involving poplar trees demonstrates the power of bioinformatics in microsatellite marker development. In this study, researchers identified over 200 microsatellite markers using bioinformatics tools like MISA and SSRLocator, some of which were later associated with disease resistance in trees (Zhao et al., 2017). The use of automated tools reduced the time and cost of marker identification and made it possible to conduct more in-depth studies of genetic traits in trees.
Advantages and Challenges of Microsatellite Marker Development
Advantages in Genetic Research
- High Polymorphism: Microsatellites exhibit a high degree of variation, making them perfect markers for studies of genetic diversity.
- Reproducibility: Once developed, microsatellite markers provide consistent results across different laboratories and populations.
- Ease of Analysis: Techniques like PCR make microsatellite analysis relatively simple and cost-effective.
Limitations and Challenges
Despite their advantages, the development of microsatellite markers comes with challenges:
- Mutation Rates: High mutation rates in microsatellites can complicate data interpretation, especially when studying closely related individuals.
- Cross-Species Amplification: Some markers may not work across different species, limiting their applicability in comparative genomics.
- Development Cost: The process of developing microsatellite markers, especially using NGS, can be costly and time-consuming.
Microsatellite Database and Tools
Popular Databases for Microsatellite Information
Several online databases provide valuable resources for microsatellite researchers:
- MITOMAP - A database focused on human mitochondrial microsatellites.
- dbSNP - Includes information on single nucleotide polymorphisms and microsatellites.
Explore more on microsatellite marker databases.
Software and Bioinformatics Tools for Marker Development
Tools such as RepeatMasker, SSRLocator, and QDD are essential for the efficient identification and analysis of microsatellite markers. For an in-depth look at various bioinformatics tools, check out our Comprehensive Guide to Microsatellite Markers.
Recent Advances and Future Directions in Microsatellite Research
Innovations in Microsatellite Marker Development
Recent technological advancements, particularly in NGS and bioinformatics, have revolutionized microsatellite marker development. The advent of CRISPR technology also offers new avenues for studying the functional roles of microsatellites in genetic diseases and evolution.
The Role of CRISPR and Other Modern Technologies
CRISPR/Cas9 gene editing, when paired with microsatellite analysis, holds the potential to address genetic diseases and explore the functional significance of microsatellites in gene expression regulation.
Industry Statistics and Market Trends in Microsatellite Research
Microsatellite marker applications are growing rapidly across various industries. For example, the global agricultural genomics market is projected to reach $10.85 billion by 2023, growing at a 12.2% CAGR. This growth is driven by the increasing adoption of molecular markers like microsatellites in crop breeding and livestock management.
| Industry Segment |
Estimated Market Value (2023) |
Expected Growth Rate (CAGR) |
| Agricultural Genomics |
$10.85 billion |
12.2% CAGR |
| Genomic Research |
$13.4 billion |
8.9% CAGR |
Learn more about our microsatellite genotyping services and how we help advance genomic research.
Conclusion
Microsatellite markers continue to play a pivotal role in genetic research, offering significant advantages in genetic mapping, biodiversity studies, and forensic science. However, challenges remain, especially in marker development and cross-species amplification. The continued advancement of technologies like NGS and CRISPR will likely drive further innovations in this field.
References:
- Weising, K., et al. (2005). Microsatellite markers: A practical guide. Plant Molecular Biology Reporter.
- Vignal, A., et al. (2002). Microsatellite-based DNA typing in animals. Animal Genetics.
- Sargent, D., & Brubaker, C. (2023). "Global Agricultural Genomics Market Size & Share Report, 2023-2030," Grand View Research.
- ResearchAndMarkets. (2022). "Genomics Research Market - Industry Trends and Forecasts," ResearchAndMarkets.


 Sample Submission Guidelines
Sample Submission Guidelines
 Distribution of microsatellite markers across the genome. (Elizabeth Bryda et al,.2008)
Distribution of microsatellite markers across the genome. (Elizabeth Bryda et al,.2008)  Schematic representation of development of microsatellite markers by cross-species amplification. (P. M. Abdul-Muneer et al,.2014)
Schematic representation of development of microsatellite markers by cross-species amplification. (P. M. Abdul-Muneer et al,.2014) 