Epitranscriptomics is a subject that studies the interaction between epigenetic modification and transcription regulation in gene expression regulation. It combines the research methods of epigenetics and transcriptomics to reveal the complex mechanism of gene expression regulation.
What is Epitranscriptomics
The field of epitranscriptomics focuses primarily on investigating chemical modifications that influence how genes are regulated, encompassing processes such as methylation of DNA, modifications to histones (including H3K27me3 and H3K4me3), and various functional aspects of non-coding RNA molecules (like miRNA and lncRNA). While these regulatory mechanisms preserve the underlying genetic sequence, they effectively control gene activity by modifying chromatin accessibility and transcription factor interactions. Epitranscriptomic research encompasses analysis of diverse RNA types found within cellular environments, organisms, and tissues—from messenger RNA to microRNA and other non-coding variants.
As a specialized domain within the broader epigenetics field, epitranscriptomics examines specific regulatory pathways affecting gene expression. The epigenetic discipline investigates heritable changes to DNA, histones, and chromatin configuration that occur without altering nucleotide sequences yet significantly impact genetic activity. By synthesizing insights from both epigenetic studies and transcriptomic analysis, epitranscriptomics elucidates the mechanisms through which modifications like DNA methylation and histone alterations regulate gene expression—specifically by controlling chromatin structure accessibility and modulating the binding capabilities of various transcription factors.
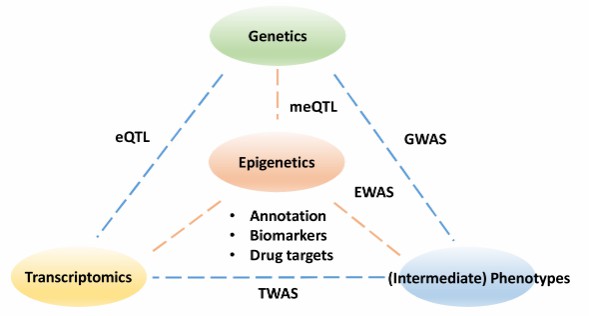 The pillars to understanding the functional impact of epigenetics (Cazaly et al., 2019)
The pillars to understanding the functional impact of epigenetics (Cazaly et al., 2019)
Epitranscriptomics plays an important role in gene expression regulation, mainly in the following aspects:
Dynamic regulation of gene expression: Epigenetic modification can dynamically regulate gene expression to adapt to environmental changes and cell functional requirements. For example, DNA methylation and histone modification play an important role in the regulation of gene expression during development.
Cell differentiation and phenotypic plasticity: Epigenetic modification plays a key role in cell differentiation and phenotypic plasticity. By changing the combination of chromatin structure and transcription factors, epigenetic modification can regulate the expression of specific genes, thus realizing the transformation of cell fate.
Study on disease mechanism: Epitranscriptomics is of great significance in the study of disease mechanism. By analyzing the epigenetic and transcriptome data of the disease, we can reveal the molecular mechanism and potential therapeutic targets of the disease. For example, epigenetic modification of miRNA plays an important role in the occurrence, development and progress of cancer.
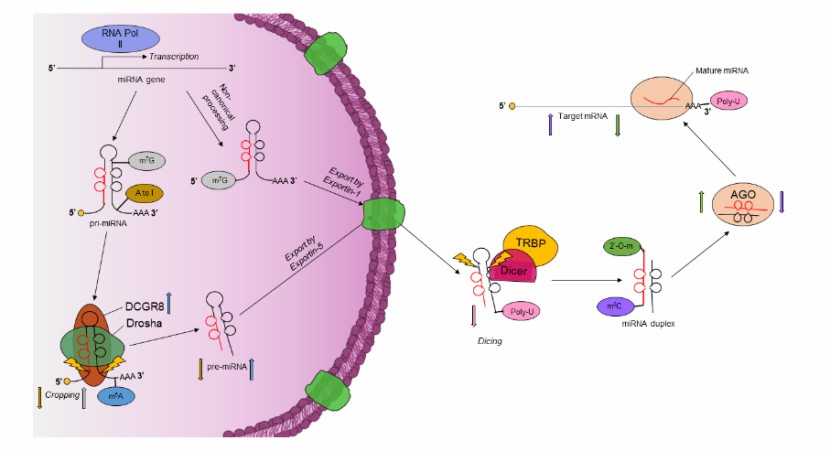 Epitranscriptomic modifications regulate the maturation and downstream targeting of miRNAs (Daniel et al., 2022)
Epitranscriptomic modifications regulate the maturation and downstream targeting of miRNAs (Daniel et al., 2022)
Multiomics data integration: Epitranscriptomics can understand the complex mechanism of gene expression regulation more comprehensively by integrating multi-omics data (such as genome, transcriptome, epigenome, etc.). For example, by integrating single cell transcriptome and epigenome data, we can reveal the dynamic changes of cell heterogeneity and gene expression regulation.
Epitranscriptomics is an interdisciplinary research field, which combines the methods of epigenetics and transcriptomics to reveal the complex mechanism of gene expression regulation. By studying the interaction between epigenetic modification and transcription regulation, epigenomics provides an important theoretical basis and practical tool for understanding organism function, disease mechanism and developing new therapeutic strategies.
Main Types of Epigenomics Modifications
Epitranscriptomics is the field of studying RNA modification, which plays an important role in RNA metabolism, function and gene expression regulation.
m6A modification
m6A (N6-methyladenosine) is one of the most common modifications in RNA, which is mainly added by methyltransferase complexes (such as METTL3 and METTL14) and removed by demethylases (such as FTO and ALKBH5). m6A modification plays a role in many aspects of RNA, including transcription, processing, transportation, translation and degradation. The dynamic change of m6A modification is realized by the interaction between methyltransferase and demethylase. This dynamic and reversible modification process makes m6A an important means to regulate RNA metabolism.
- Transcription: m6A modification affects the splicing, maturation and nucleation of RNA.
- Processing: m6A modification is involved in the secondary structure formation and stability regulation of RNA.
- Transport: m6A modification affects the transport and localization of RNA in cells.
- Translation: m6A modification regulates the translation efficiency and selectivity of mRNA through the recognition of YTHDF family proteins.
- Degradation: m6A modification can promote or inhibit the degradation of RNA.
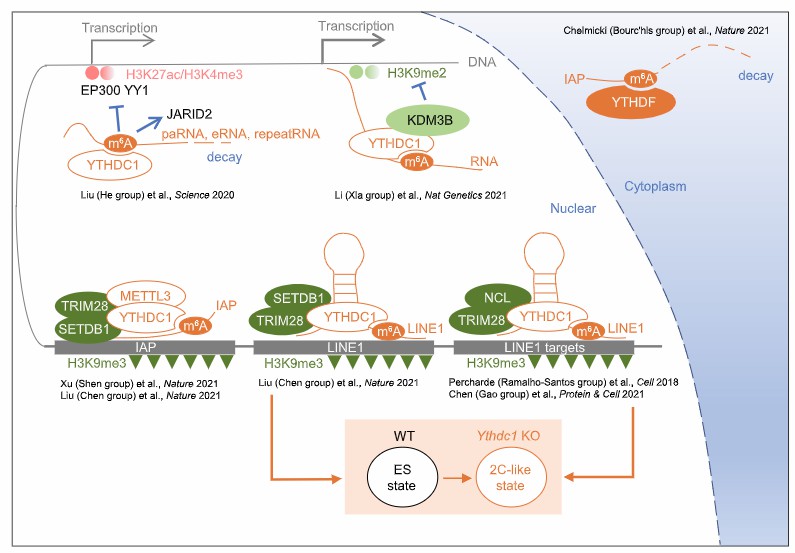 Schematic of m6A regulating chromatin environment (He et al., 2021)
Schematic of m6A regulating chromatin environment (He et al., 2021)
m6A modification regulates gene expression through various mechanisms:
- Direct regulation: m6A modification can affect the translation efficiency and stability of mRNA, thus directly regulating gene expression.
- Indirect regulation: m6A modification indirectly affects gene expression by affecting RNA splicing, processing and transportation.
- Synergism with other modifications: m6A modification interacts with other epigenetic modifications (such as histone H3K9me3) to jointly regulate gene expression.
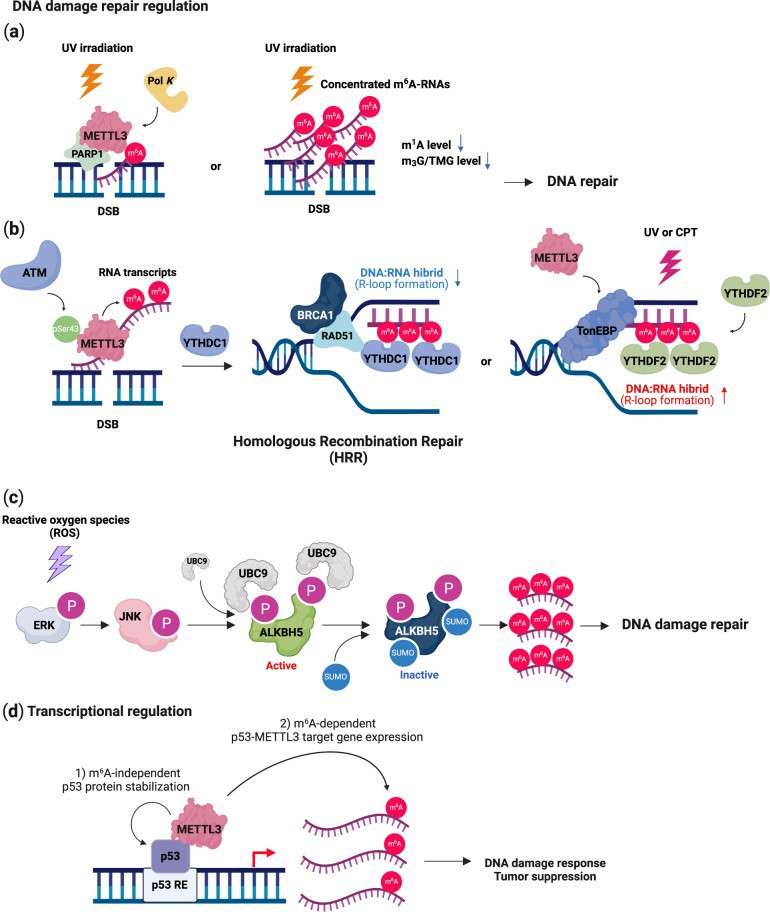 DNA damage response and repair by m6A (Hong et al., 2022)
DNA damage response and repair by m6A (Hong et al., 2022)
Other RNA modifications
Besides m6A modification, other RNA modifications such as pseudouridine and 5-methylcytosine (m5C) also play an important role in RNA metabolism:
- Pseudouridine modification mainly occurs in rRNA, which affects protein synthesis by improving the stability and translation efficiency of rRNA.
- m5C modification mainly occurs in tRNA and rRNA, and regulates gene expression and cell metabolism by affecting the structure and function of RNA.
Relationship between RNA modification and diseases
- Neurodegenerative diseases: m6A modification plays a key role in neurodegenerative diseases such as Alzheimer's disease and Parkinson's disease, by regulating neuron function and cell fate.
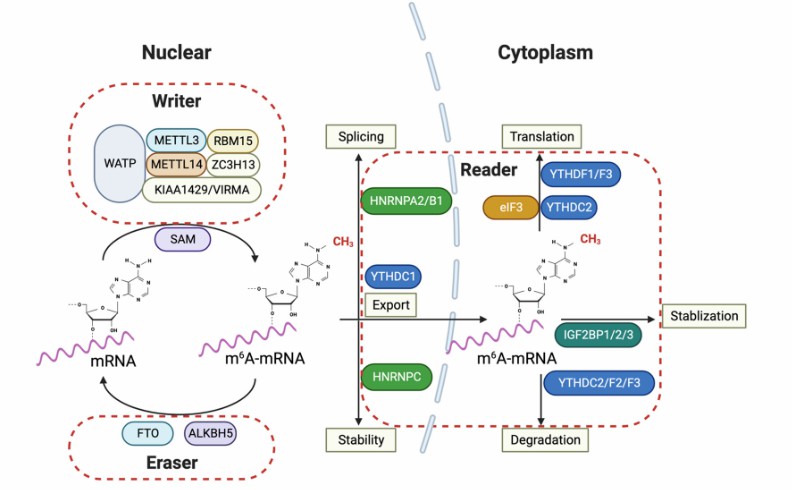 m6A RNA modification and its regulatory proteins in glioma pathogenesis (Kobayashi et al., 2024)
m6A RNA modification and its regulatory proteins in glioma pathogenesis (Kobayashi et al., 2024)
- Tumors: m6A modification plays an important role in the occurrence and development of tumors, and promotes the proliferation and metastasis of tumor cells by regulating gene expression and cell cycle.
- Metabolic diseases: m6A modification plays an important role in metabolic diseases such as obesity and diabetes, and affects the occurrence and development of diseases by regulating lipid metabolism and glucose metabolism.
- Immune system diseases: m6A modification plays an important role in immune system diseases, and affects immune response by regulating the function of immune cells and the expression of cytokines.
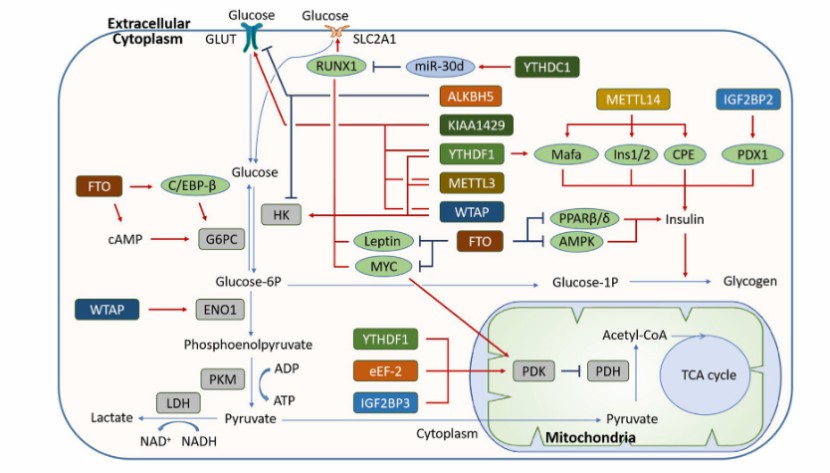 The function of m6A modification in carbohydrate metabolism (Hu et al., 2023)
The function of m6A modification in carbohydrate metabolism (Hu et al., 2023)
- Pseudouridine: It plays an important role in virus replication and host cell metabolism, and regulates virus replication by affecting the stability and translation efficiency of viral RNA.
- m5C: It plays an important role in cardiovascular diseases, and affects vascular function and cardiovascular health by regulating gene expression and cell metabolism.
m6A modification and other RNA modifications in epigenomics play an important role in RNA metabolism, gene expression regulation and the occurrence and development of many diseases. Future research will further reveal the specific mechanism of these modifications and their potential applications in disease treatment.
Services you may interested in
Application of Epitranscriptomics in Diseases
The application of epigenomics in diseases has broad research prospects and practical significance, especially in the fields of cancer research, nervous system diseases and personalized medicine.
Application of Epitranscriptomics in Cancer Research
The application of epigenomics in cancer research mainly focuses on the following aspects:
- Cancer diagnosis and treatment decision: Whole genome and transcriptome sequencing technology can provide more comprehensive tumor information, including mutation characteristics, gene fusion, transcriptional regulation and epigenetic changes. This information helps doctors identify treatable variations and guide individualized treatment programs. For example, in childhood and young adult cancers, adult B-lymphoma, acute myeloid leukemia and acute lymphoblastic leukemia, the application of epigenomics has shown remarkable clinical value.
- Precision oncology: Radiotranscriptomics technology captures the transcription state of cells through RNA-mediated gene expression regulation, and reveals patient-specific epigenetic changes and environmental changes. These changes are related to the acquisition of pathogenicity and can be used for the diagnosis of early diseases or multifactorial diseases.
- Multiomics integration analysis: By integrating genome, transcriptome, epigenome and protein data, researchers can better understand the disease mechanism. For example, in the study of hepatocellular carcinoma (HCC), integration analysis reveals the chromatin structure and active chromatin regions related to gene expression level, which provides new molecular targets and drug markers for the treatment of HCC.
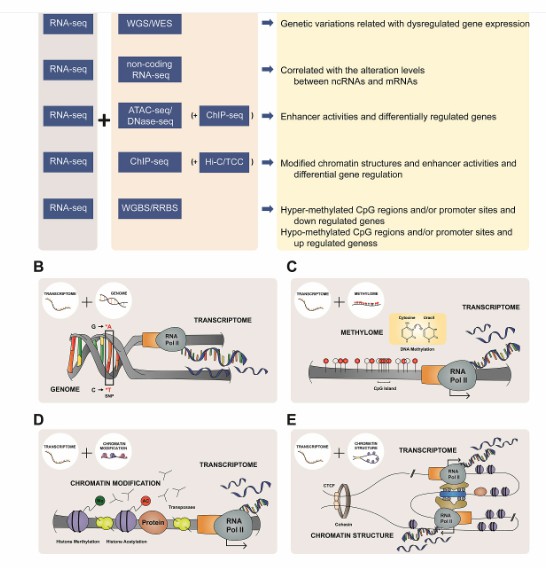 Integrative analysis with complex biological meanings (Oh et al., 2020)
Integrative analysis with complex biological meanings (Oh et al., 2020)
Association between RNA modification and nervous system diseases
The role of RNA modification in nervous system diseases has attracted more and more attention.
- RNA modification and neurodegenerative diseases: RNA modification, especially m6A modification, has an important influence on the stability, localization, splicing and translation of RNA. These modifications play a key role in nervous system diseases. For example, the role of m6A modification in Alzheimer's disease (AD) has been widely studied. By integrating multiple omics data, researchers constructed a co-expression network and a dependency network, and further explored the role of mitochondrial-related nuclear genes in AD.
- RNA modification and scleroderma: The mechanism of RNA m6A modification in scleroderma has also made progress. Through the mouse model induced by bleomycin, the researchers observed the change of m6A modification and discussed its potential mechanism.
The potential of epigenomics in personalized medicine
The application of epigenomics in personalized medicine is mainly reflected in the following aspects:
- Precision medicine: Epitranscriptomics, combined with genomics, protein genomics and metabonomics, can provide personalized treatment for patients. For example, by integrating multiple omics data, researchers can build accurate disease models and identify potential therapeutic targets.
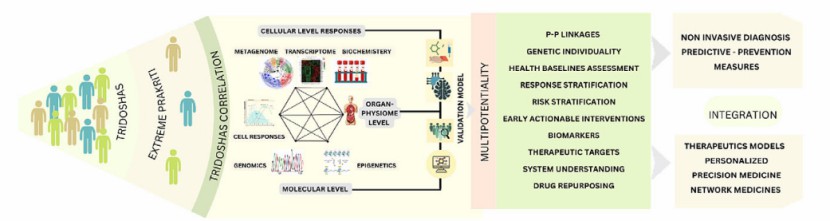 Iterative approach of Ayurgenomcs for the development of an operational framework of network medicine based on concepts of Ayurveda (Mukerji et al., 2023)
Iterative approach of Ayurgenomcs for the development of an operational framework of network medicine based on concepts of Ayurveda (Mukerji et al., 2023)
- Drug development: The application of epigenetics in drug development also shows great potential. By analyzing protein epigenetic biomarkers, researchers can better understand the diagnosis and treatment of brain diseases.
- Big data analysis: The application of big data analysis in the field of health care promotes the informed diagnosis, treatment and disease prevention of patients. By tapping the data potential, big data analysis improves the efficiency of medical organizations.
- Application of AI technology: The application of artificial intelligence (AI) technology in the field of bioinformatics has accelerated the research process and enhanced the ability of data interpretation. AI tools and algorithms are promoting the innovation of biomedical science, and will play an important role in integrating a variety of multi-disciplinary data in the future.
Epitranscriptomics has a broad application prospect in cancer research, nervous system diseases and personalized medicine. Through the application of multi-omics integration analysis, single cell multi-omics technology and AI technology, researchers can better understand the disease mechanism and provide new ideas and methods for disease prevention, diagnosis and treatment.
Detection Approaches of Epitranscriptomics
Detection of RNA modification is an important basis for epigenetics research. At present, the commonly used detection methods of RNA modification include:
- Immunoprecipitation combined with sequencing (MeRIP-seq): This is one of the most commonly used methods. The target RNA is modified by immunoprecipitation with specific antibodies, and then high-throughput sequencing is performed. For example, the m6A modification can be immunoprecipitated by m6A antibody and then sequenced.
- Liquid chromatography-mass spectrometry (LC-MS/MS): This method can quantify and identify all RNA modifications and is suitable for the analysis of complex samples.
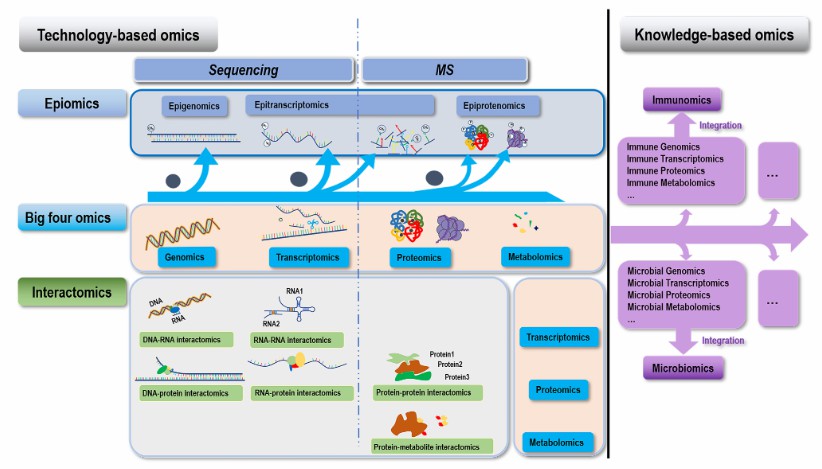 RNA modification combined with MS (Dai et al., 2022)
RNA modification combined with MS (Dai et al., 2022)
- High-throughput sequencing technology: For example, RNA sequencing (RNA-seq) can comprehensively analyze the distribution and function of RNA modification.
- ELISA: Although ELISA is mainly used for the detection of protein modification, it can also be used for the quantitative analysis of some RNA modifications, such as the detection of m6A modification.
Sequencing technology and analysis method based on RNA modification
With the development of sequencing technology, the detection methods of RNA modification are also improving:
Direct high-throughput sequencing: Such as MeRIP-seq, m6A-seq, etc. These methods directly capture the target RNA modification through specific antibodies or chemical markers, and then carry out high-throughput sequencing.
Auxiliary sequencing method: for example, chemical labeling (such as methylation-specific probe) or enzyme-assisted method (such as ADAR) is used to transform RNA modification into a measurable signal through chemical or enzymatic reaction.
Metabolic marker sequencing: RNA modification is transformed into a sequence-measurable fragment by metabolic marker technology (such as hypoxanthine-guanine phosphoribosyl transfer, HGPRT).
Nanopore Direct RNA Sequencing: RNA modification can be directly detected by nano-pore sequencing technology without complicated chemical or enzymatic steps.
Future Directions and Challenges of Epigenomics
Epitranscriptomics reveals the important role of RNA modification in gene expression regulation through various high-throughput sequencing techniques and analysis methods. Future research will pay more attention to multi-dimensional analysis, single cell level research and clinical application, and develop more efficient new technologies to deal with complex biological problems.
In the research direction, technological innovation will be an important force to promote the development of this field. The technology of single cell epigenomics will continue to improve. Although the RNA modification of a single cell can be analyzed at present, there is still room for improvement in resolution and flux. In the future, it is expected to achieve more accurate mapping of single cells, so as to deeply analyze the dynamic changes of RNA modification in different cell subsets.
In addition, the technology of spatial epigenome will also usher in a breakthrough, which can determine the spatial distribution of RNA modification in tissue microenvironment, which is very important to reveal the regulatory mechanism of epigenome in intercellular communication and tissue function maintenance. For example, in the study of tumor microenvironment, this technology can be used to explore the interaction of RNA modification between immune cells and tumor cells.
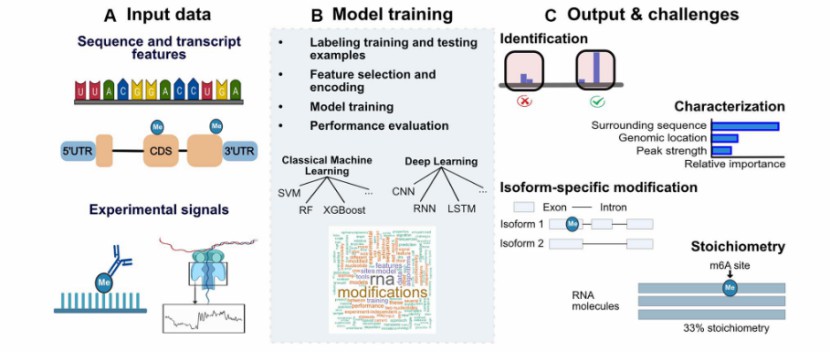 Transcriptome-wide prediction of chemical messenger RNA modifications with ML (Acera et al., 2023)
Transcriptome-wide prediction of chemical messenger RNA modifications with ML (Acera et al., 2023)
In terms of functional mechanism, the discovery and functional analysis of new RNA modifications will become the focus. In addition to the well-known modifications such as m6A and m5C, it is expected that there will be more modifications that have not been discovered before. By integrating the data of transcriptome, protein group and metabolomics, we can fully analyze the role of new RNA modification in gene expression regulation, cell fate determination and disease occurrence and development. However, the development of epigenomics also faces many challenges. From a technical point of view, the accuracy, sensitivity and flux of the current detection technology are still difficult to reach the ideal state, and the comparability of data between different technical platforms needs to be improved.
In the study of biological mechanism, although some functions of RNA modification have been found, the specific mechanism of most modifications is still poorly understood, and the complex interaction network between RNA modification and other biomolecules needs to be further explored. In addition, in clinical application transformation, how to effectively apply basic research results to disease diagnosis, treatment and prognosis evaluation is also a difficult problem to be overcome.
In the future, the study of epigenomics needs interdisciplinary cooperation, continuous optimization of technical means and in-depth exploration of biological mechanisms to meet these challenges, promote the continuous development of this field and bring more breakthroughs to the fields of life science and medicine.
References:
- Cazaly E, Saad J., et al. "Making Sense of the Epigenome Using Data Integration Approaches." Front Pharmacol. 2019 10: 126 https://doi.org/10.3389/fphar.2019.00126
- Del Valle-Morales D, Le P., et a;. "The Epitranscriptome in miRNAs: Crosstalk, Detection, and Function in Cancer." Genes (Basel). 2022 13(7):1289 https://doi.org/10.3390/genes13071289
- He C, Lan F. "RNA m6A meets transposable elements and chromatin." Protein Cell. 2021 12(12):906-910 https://doi.org/10.1007/s13238-021-00859-2
- Hong J, Xu K, Lee JH. "Biological roles of the RNA m6A modification and its implications in cancer." Exp Mol Med. 2022 54(11):1822-1832 https://doi.org/10.1038/s12276-022-00897-8
- Kobayashi A, Kitagawa Y., et al. "Emerging Roles and Mechanisms of RNA Modifications in Neurodegenerative Diseases and Glioma." Cells. 2024 13(5):457 https://doi.org/10.3390/cells13050457
- Hu H, Li Z., et al. "Insights into the role of RNA m6A modification in the metabolic process and related diseases." Genes Dis. 2023 11(4):101011 https://doi.org/10.1016/j.gendis.2023.04.038
- Oh S, Jo Y., et al. "From genome sequencing to the discovery of potential biomarkers in liver disease." BMB Rep. 2020 53(6):299-310 https://doi.org/10.5483/bmbrep.2020.53.6.074
- Mukerji M. "Ayurgenomics-based frameworks in precision and integrative medicine: Translational opportunities." Camb Prism Precis Med. 2023 1:e29 https://doi.org/10.1017/pcm.2023.15
- Dai X, Shen L. "Advances and Trends in Omics Technology Development." Front Med (Lausanne). 2022 9:911861 https://doi.org/10.3389/fmed.2022.911861
- Acera Mateos P, Zhou Y, Zarnack K, Eyras E. "Concepts and methods for transcriptome-wide prediction of chemical messenger RNA modifications with machine learning." Brief Bioinform. 2023 24(3):bbad163 https://doi.org/10.1093/bib/bbad163


 Sample Submission Guidelines
Sample Submission Guidelines
 The pillars to understanding the functional impact of epigenetics (Cazaly et al., 2019)
The pillars to understanding the functional impact of epigenetics (Cazaly et al., 2019) Epitranscriptomic modifications regulate the maturation and downstream targeting of miRNAs (Daniel et al., 2022)
Epitranscriptomic modifications regulate the maturation and downstream targeting of miRNAs (Daniel et al., 2022) Schematic of m6A regulating chromatin environment (He et al., 2021)
Schematic of m6A regulating chromatin environment (He et al., 2021) DNA damage response and repair by m6A (Hong et al., 2022)
DNA damage response and repair by m6A (Hong et al., 2022) m6A RNA modification and its regulatory proteins in glioma pathogenesis (Kobayashi et al., 2024)
m6A RNA modification and its regulatory proteins in glioma pathogenesis (Kobayashi et al., 2024) The function of m6A modification in carbohydrate metabolism (Hu et al., 2023)
The function of m6A modification in carbohydrate metabolism (Hu et al., 2023) Integrative analysis with complex biological meanings (Oh et al., 2020)
Integrative analysis with complex biological meanings (Oh et al., 2020) Iterative approach of Ayurgenomcs for the development of an operational framework of network medicine based on concepts of Ayurveda (Mukerji et al., 2023)
Iterative approach of Ayurgenomcs for the development of an operational framework of network medicine based on concepts of Ayurveda (Mukerji et al., 2023) RNA modification combined with MS (Dai et al., 2022)
RNA modification combined with MS (Dai et al., 2022) Transcriptome-wide prediction of chemical messenger RNA modifications with ML (Acera et al., 2023)
Transcriptome-wide prediction of chemical messenger RNA modifications with ML (Acera et al., 2023)