CD Genomics offers state-of-the-art Nanopore Direct RNA Sequencing services for accurate detection of RNA modifications. Our platform provides high-throughput analysis, delivering detailed insights into RNA structure and function. This service supports research in gene expression, transcriptomics, and molecular biology studies.
The Introduction of Nanopore Direct RNA Sequencing
Current the high-throughput sequencing of complementary DNA (cDNA) (NGS-RNA-seq) have enabled transcriptome-wide studies of RNA biology, These RNA-seq methods detect the products of a synthesis reaction rather than directly detecting the RNA molecule. New third-generation sequencing are being developed to overcome limitations such as amplification biases, lack of single-molecule sensitivity and isoform ambiguity. This improves the ease and speed of RNA analysis, while yielding richer biological information. Furthermore, NGS-RNA-seq is not particularly suitable for the analysis of short, degraded and/or small quantity RNA samples.
Here we CD Genomics provide nanopore direct single molecule RNA sequencing, a highly parallel, real-time, single-molecule method that without prior conversion of RNA to cDNA. This method yields full-length, strand-specific RNA sequences and enables the direct detection of nucleotide analogs in RNA, and provides a path to high-throughput and low-cost direct RNA sequencing, achieves the ultimate goal of a comprehensive and bias-free understanding of transcriptomes.
Oxford Nanopore Technologies' nanopore-based platform detects single molecules of DNA, RNA, proteins and small molecules as they traverse through a nanopore, without the need for an enzymatic synthesis reaction. The platform consists of single nanopores embedded in an array of thousands of individual synthetic polymer membranes on a single flowcell. An electric potential drives DNA toward and into the nanopores. When a single DNA molecule is captured in a pore and ratcheted through the pore at a consistent rate by an engineered motor protein, it creates perturbations of the nanopore current which a recurrent neural network (RNN) converts into base sequences. This technology offers long sequencing reads (up to 2Mb) and detection of epigenetic markers.

Principle of Nanopore Direct RNA Sequencing
In nanopore sequencing, the motor proteins that sequence RNA strands progressively move along the native RNA strand. These enzymes operate in the opposite direction to DNA helicases, resulting in RNA strands translocating through the nanopore in the 3'–5' direction, contrasting with the 5'-3' direction of DNA signals. The motor protein in RNA sequencing adapters is specifically designed to control RNA passage through the nanopore, regulating RNA translocation at an average speed of 70 nucleotides per second. Compared to motor proteins used in cDNA-based sequencing, those employed in direct RNA sequencing significantly reduce speed, resulting in a notable decrease in throughput.
Key Features and Advantages of Nanopore Direct RNA Sequencing
- Low input amounts combined with rapid, streamlined workflows enable highly sensitive gene expression analysis.
- Full-length transcripts-The high yields of long, full-length reads delivered by nanopore sequencing allow unambiguous identification of splice variants and gene fusions
- Accurate transcript and isoform quantification
- Eliminate PCR bias using direct RNA sequencing
- Detect base modifications alongside nucleotide sequence using direct RNA
- Easy identification of anti-sense transcripts
- Accurately detect transcript poly(A) tail lengths.
Applications of Nanopore Direct RNA Sequencing
- Gene function: Focus on samples carrying specific functions to reveal the main causes of different functions
- Gene structure: Alternative splicing, APA, fusion gene, SSR, CDS prediction, TSS/TES identification, etc.
- Full-length transcript quantification: Find comprehensive and effective differential transcripts and identify functional genes
- RNA methylation: Direct full-length transcriptome sequencing can detect base modifications at the RNA level, such as m6A/m5C
- Virus RNA research
- Sequencing native RNA
- Exploring RNA modifications
Nanopore Direct RNA Sequencing Workflow
Direct RNA sequencing was performed using the nanopore sequencing platform, with 500-700 ng of poly(A) RNA (to ensure sufficient material remained for sequencing) used as input for library preparation. The RNA was ligated to a poly(T) adaptor using T4 DNA ligase. Following adaptor ligation, the products were purified by RNAClean beads. Sequencing adaptors preloaded with motor protein were then ligated onto the overhang of the previous adaptor using T4 DNA ligase, and the final library was cleaned up once more. The RNA library was quantified using a Qubit fluorometer. The final RNA libraries were added to flowcells and carried out on the nanopore sequencing platform.
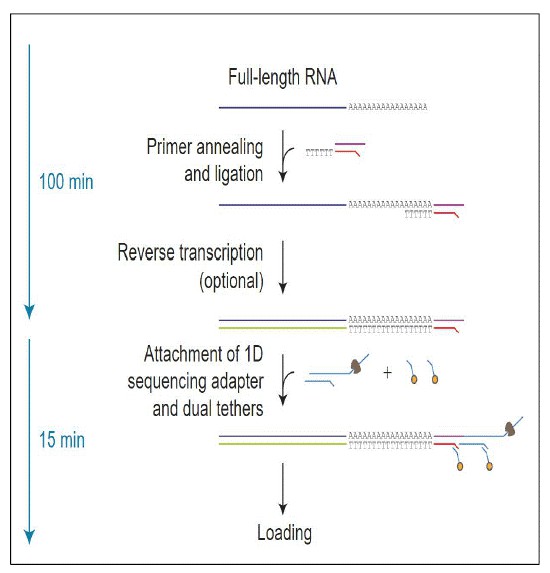 Figure 1. Library preparation steps for direct RNA sequencing.
Figure 1. Library preparation steps for direct RNA sequencing.

Service Specifications
Sample Requirements
|
|
Click |
Sequencing Strategy
|
| Bioinformatics Analysis
We provide multiple customized bioinformatics analyses:
|
Analysis Pipeline
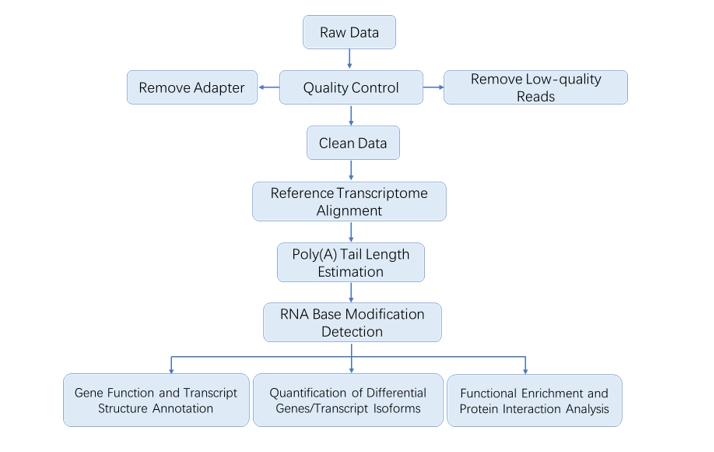
Deliverables
- The original sequencing data
- Experimental results
- Data analysis report
- Details in Nanopore Direct RNA Sequencing for your writing (customization)
Discover RNA modification capabilities with CD Genomics' Nanopore Direct RNA Sequencing service. We provide direct sequencing of RNA along with thorough bioinformatics analysis to reveal detailed genetic information. Contact us to elevate your research endeavors.
References
- Garalde D R , Snell E A , Jachimowicz D , et al. Highly parallel direct RNA sequencing on an array of nanopores. Nature Methods, 2018, 15(3).
- A I S , A K A C D S , C J M B , et al. Evaluating the potential of direct RNA nanopore sequencing: Metatranscriptomics highlights possible seasonal differences in a marine pelagic crustacean zooplankton community. Marine Environmental Research, 153.
- Feng, Jiang, Jie, et al. Long-read direct RNA sequencing by 5'-Cap capturing reveals the impact of Piwi on the widespread exonization of transposable elements in locusts. Rna Biology, 2019.
- Lorenz D A , Sathe S , Einstein J M , et al. Direct RNA sequencing enables m6A detection in endogenous transcript isoforms at base specific resolution. RNA, 2019, 26(1): rna.072785.119.
Partial results are shown below:
1. Why is direct RNA sequencing not recommended for gene expression, splicing isoform, and fusion protein analysis?
One reason is the current lower throughput compared to cDNA sequencing, resulting in relatively higher costs for generating sufficient data. As cDNA sequencing can adequately address these issues, direct RNA nanopore sequencing is a novel and evolving technology. Subsequent advancements by ONT company are expected to enhance the throughput of direct RNA sequencing significantly, promising substantial increases in yield.
2. How does Nanopore Direct RNA Sequencing differ from traditional RNA sequencing methods?
Traditional RNA sequencing methods typically involve reverse transcription of RNA into cDNA followed by PCR amplification. Nanopore Direct RNA Sequencing, on the other hand, directly reads RNA molecules through nanopores, avoiding reverse transcription and PCR steps, thereby providing more direct and comprehensive transcript information.
3. What are the main challenges of Nanopore Direct RNA Sequencing?
The primary challenges of Nanopore Direct RNA Sequencing encompass several key aspects:
- Addressing high single-read error rates necessitates the development of advanced error correction algorithms to ensure data accuracy.
- Managing extensive data processing and storage demands, predominantly attributed to the elongated read lengths characteristic of this sequencing method.
- Overcoming the intricacies associated with technical procedures and data interpretation, which mandate specialized expertise and meticulously optimized experimental settings for successful implementation and analysis.
Quantitative profiling of N6-methyladenosine at single-base resolution in stem-differentiating xylem of Populus trichocarpa using Nanopore direct RNA sequencing
Journal: Genome biology
Impact factor: 13.6
Published: 07 January 2021
Background
Recent advancements in RNA modification research emphasize the significance of N6-Methyladenosine (m6A) in plant gene regulation. Techniques like MeRIP-Seq/m6A-seq identify m6A-enriched transcripts in Arabidopsis thaliana, but UV crosslinking efficiency limits plant studies. New methods such as m6A-REF-seq and MAZTER-seq offer quantitative profiling of m6A sites but focus on ACA motifs, missing others like DRACH. Oxford Nanopore's direct RNA sequencing (DRS) shows promise in detecting RNA modifications. Methods like EpiNano and MINES provide qualitative m6A profiles using DRS, yet distinguishing m6A from other modifications remains a challenge. Ionic current-based models improve m6A identification, supporting single-base resolution studies for comparing m6A across samples.
Materials & Methods
Sample Preparation
- Populus trichocarpa (P. trichocarpa)
- Stem
- Total RNA extraction
Sequencing
- Construction of library
- Nanopore Direct RNA Sequencing
- MeRIP-Seq
- m6A-REF-seq
- Alignment with the reference genome
- GO enrichment analysis
- Measurement of poly(A) tail length
Results
This study on m6A in Populus trichocarpa's stem-differentiating xylem identified major m6A components and utilized Nanopore direct RNA sequencing (DRS) with GridION and MinION platforms. Basecalling via Guppy and alignment with minimap2 enabled m6A prediction using a trained XGBoost model. The analysis revealed enriched m6A near stop codons and 3'UTRs, supported by high reproducibility between biological repeats. The study found m6A enrichment in genes involved in wood formation, suggesting a potential role in regulating gene expression stability and levels in plant tissues. Comparisons with EpiNano and MINES highlighted Nanom6A's advantage in detecting low m6A/A ratios and single-transcript resolution.
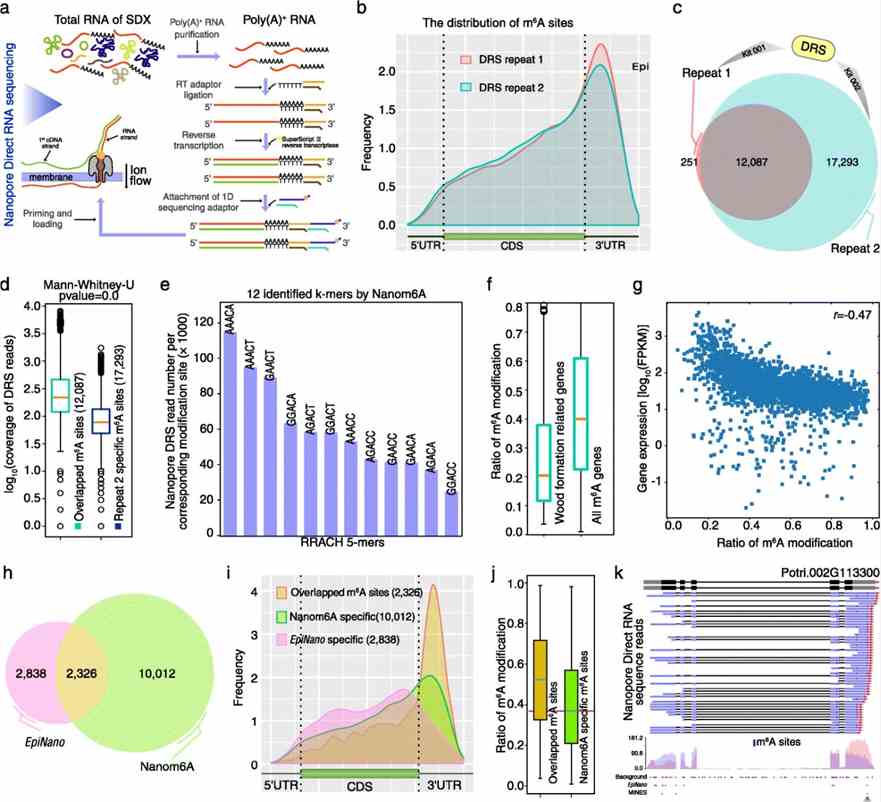 Fig 1. Quantitative profiling of m6A in stem-differentiating xylem of Populus trichocarpa.
Fig 1. Quantitative profiling of m6A in stem-differentiating xylem of Populus trichocarpa.
To validate Nanom6A predictions, MeRIP-Seq with an anti-m6A antibody confirmed m6A enrichment near stop codons and 3'UTRs, with higher coverage from deeper sequencing suggesting detection of low abundance transcripts. MeRIP-Seq detected 81% of predicted m6A-modified genes and covered 49% of Nanopore DRS-predicted m6A sites at single-base resolution. Additionally, m6A-REF-seq validated 80% of Nanom6A-detected m6A sites, confirming the reliability of direct RNA sequencing for m6A identification.
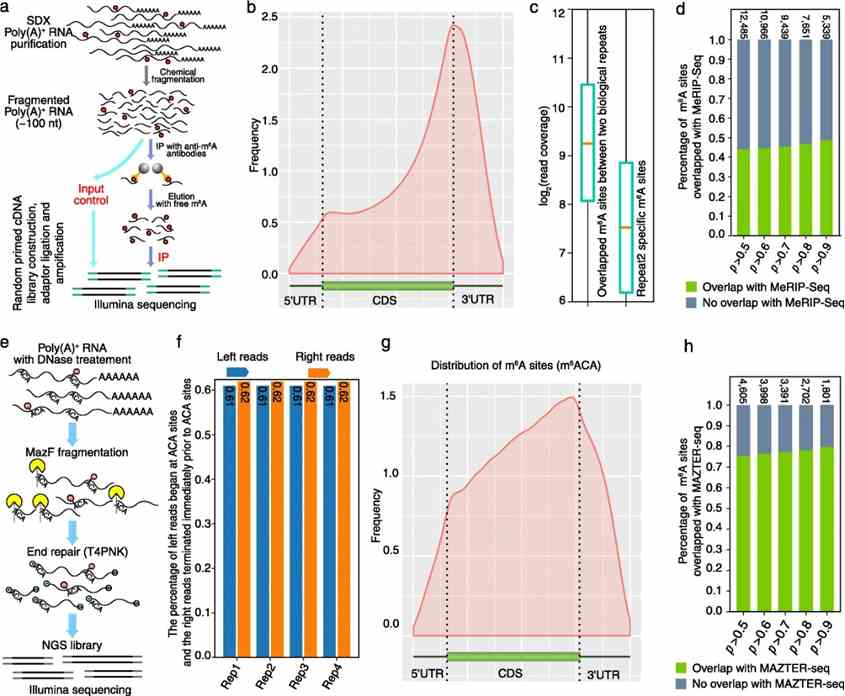 Fig 2. Validation of Nanom6A using MeRIP-Seq and m6A-REF-seq/MAZTER-seq.
Fig 2. Validation of Nanom6A using MeRIP-Seq and m6A-REF-seq/MAZTER-seq.
In this study of Populus trichocarpa's stem-differentiating xylem, Nanopore DRS efficiently detected poly(A) tail lengths and quantified m6A modifications at single-base resolution. Poly(A) tail lengths averaged 92 nt (mean) and 82 nt (median), showing a negative correlation with gene expression. Alternative polyadenylation analysis revealed 2421 genes with significant differences in poly(A) length between proximal and distal sites, impacting gene regulation in processes like secondary wall biosynthesis. Additionally, m6A quantification highlighted differential modification patterns between transcripts with proximal and distal poly(A) sites, suggesting potential regulatory roles in mRNA stability and translation.
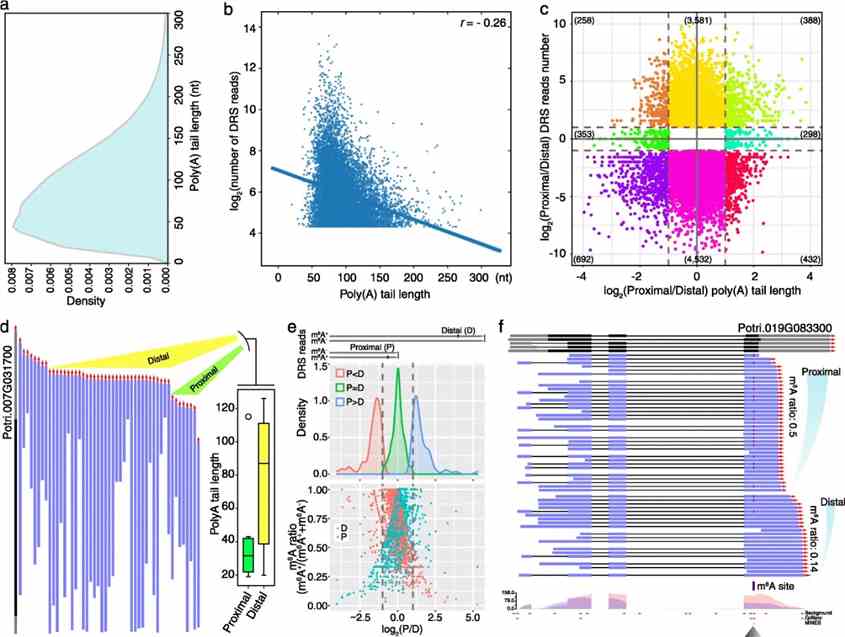 Fig 3. The differential length of poly(A) tail and differential m6A modification on alternative polyadenylation sites.
Fig 3. The differential length of poly(A) tail and differential m6A modification on alternative polyadenylation sites.
Conclusion
Nanopore direct RNA sequencing detects RNA base modifications like m6A, distinct from methods like EpiNano and MINES. Nanom6A, using an XGBoost model, achieves single-base resolution for m6A identification and quantification at transcript level. Validation with MeRIP-Seq and m6A-REF-seq confirms Nanom6A's accuracy, expanding Nanopore DRS applications in studying m6A regulatory mechanisms in SDX and alternative polyadenylation.
Reference
- Gao Y, Liu X, Wu B, et al. Quantitative profiling of N6-methyladenosine at single-base resolution in stem-differentiating xylem of Populus trichocarpa using Nanopore direct RNA sequencing. Genome biology. 2021, 22:1-7.
Here are some publications that have been successfully published using our services or other related services:
FIONA1-mediated methylation of the 3'UTR of FLC affects FLC transcript levels and flowering in Arabidopsis
Journal: PLoS Genetics
Year: 2022
The m6A writer FIONA1 methylates the 3'UTR of FLC and controls flowering in Arabidopsis
Journal: bioRxiv
Year: 2022
Complete Genome Sequence of the Lignocellulose-Degrading Actinomycete Streptomyces albus CAS922
Journal: Microbiology Resource Announcements
Year: 2020
See more articles published by our clients.


 Sample Submission Guidelines
Sample Submission Guidelines
