The field of genomics has undergone dramatic transformations in recent years, enabling researchers to gain deeper insights into the complexities of gene expression. Among the most impactful innovations are bulk RNA sequencing and single-cell RNA sequencing. While both are powerful tools for exploring gene activity, they offer different advantages depending on the research goals. This article will compare these two methods, highlighting their differences, applications, challenges, and more, helping you choose the right approach for your next project.
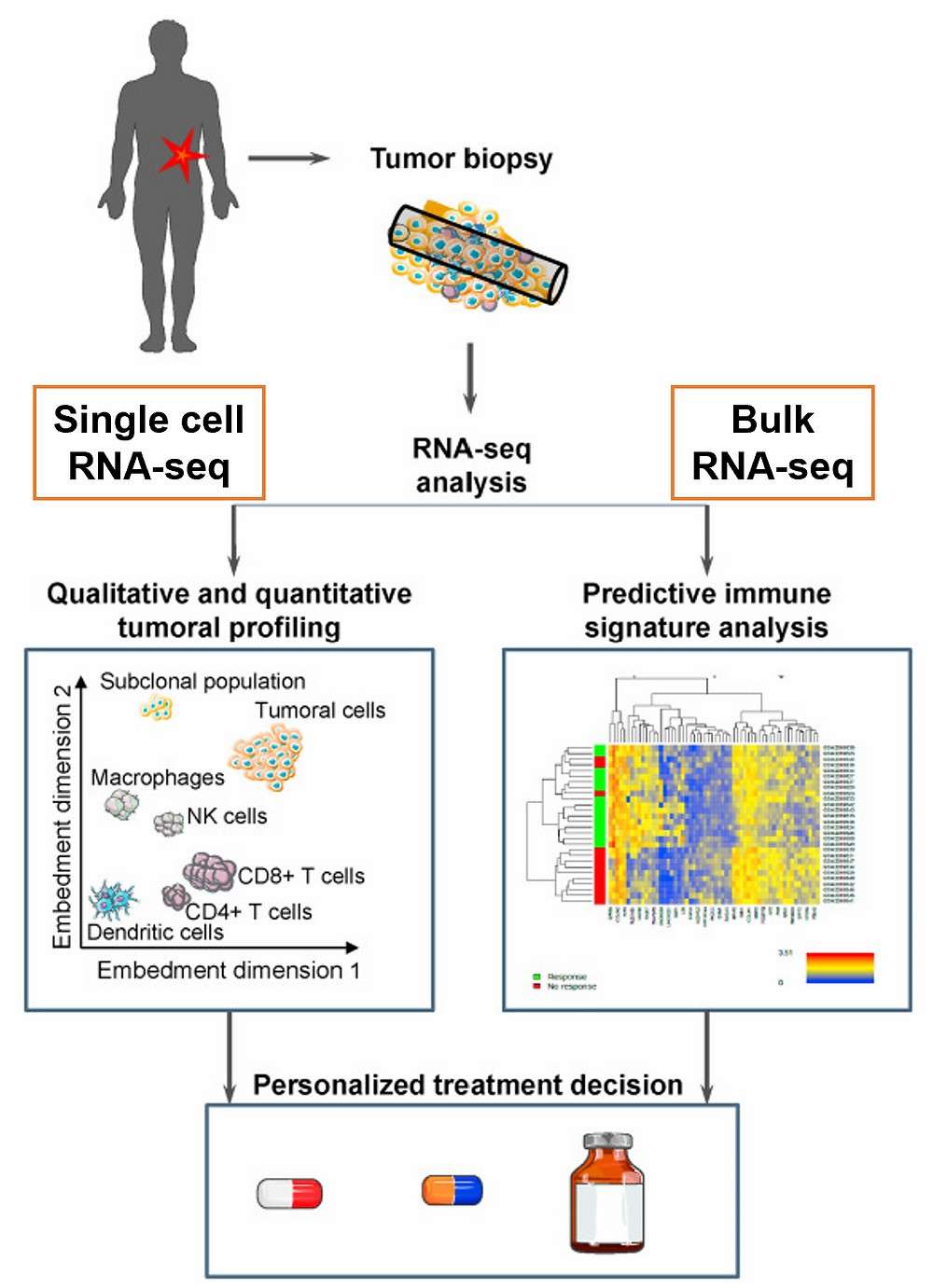 Figure 1. Applications of single-cell and bulk RNA sequencing in onco-immunology(Maria Kuksin et al,. 2021)
Figure 1. Applications of single-cell and bulk RNA sequencing in onco-immunology(Maria Kuksin et al,. 2021)
1. Understanding Bulk RNA Sequencing
Bulk RNA sequencing (RNA-Seq) is a method used to analyze gene expression from a population of cells, typically collected from tissues or cell cultures. Here's how it works:
- How it Works: Bulk RNA sequencing provides a comprehensive gene expression profile for a group of cells. The RNA is extracted, converted to complementary DNA (cDNA), and sequenced to quantify gene expression levels across the entire sample.
- Advantages:
- Lower cost: Bulk RNA sequencing is significantly more affordable than single-cell sequencing.
- Simpler data analysis: The data is easier to process because it represents an average gene expression across the entire population.
- Ideal for homogeneous samples: It works best when studying tissues or cell populations with similar characteristics.
If you want to explore the different sequencing platforms and methods, our Select Platform for RNA Sequencing page provides additional details.
2. Exploring Single-Cell RNA Sequencing
Single-cell RNA sequencing (scRNA-Seq) takes gene expression analysis to a whole new level by examining gene activity at the individual cell level.
- How it Works: Unlike bulk RNA sequencing, scRNA-Seq isolates individual cells before sequencing. Each cell's RNA is analyzed separately, allowing researchers to investigate gene expression variations within a heterogeneous population.
- Advantages:
- High resolution: Single-cell RNA sequencing reveals gene expression at a single-cell resolution, providing a detailed map of cellular activity.
- Cellular heterogeneity detection: It helps identify rare cell types or subpopulations that bulk sequencing may miss.
- Ideal for complex tissues: scRNA-Seq excels in understanding the diversity of cells within complex tissues, such as tumors or immune cell populations.
For more information on single-cell RNA sequencing, visit our Single-cell RNA Sequencing: Introduction, Methods, and Applications page.
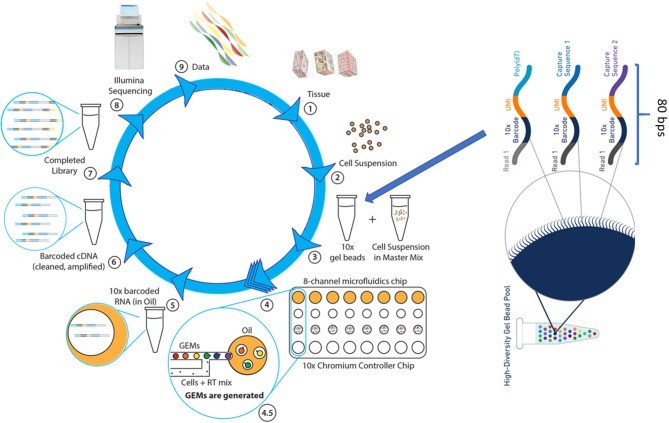 Figure 2. The single cell RNASeq. (Ye Wang et al,.2020)
Figure 2. The single cell RNASeq. (Ye Wang et al,.2020)
3. Key Differences Between Bulk and Single-Cell Sequencing
Here's a comparison of the key features of bulk RNA sequencing and single-cell RNA sequencing:
| Feature |
Bulk RNA Sequencing |
Single-Cell RNA Sequencing |
Examples |
| Resolution |
Average of cell population |
Individual cell level |
Patel et al. (2014) on glioblastoma demonstrated that single-cell RNA-seq revealed intratumoral heterogeneity and identified clinically relevant subpopulations, which was not possible with bulk sequencing1. |
| Cost |
Lower (~1/10th of scRNA-seq) |
Higher |
Stark et al. (2019) reported that bulk RNA-seq costs were approximately $300 per sample, while single-cell RNA-seq ranged from $500 to $2000 per sample depending on the platform2. |
| Data Complexity |
Lower |
Higher |
Lähnemann et al. (2020) highlighted that single-cell RNA-seq data analysis requires specialized computational methods due to increased noise and sparsity compared to bulk RNA-seq3. |
| Cell Heterogeneity Detection |
Limited |
High |
Villani et al. (2017) used single-cell RNA-seq to identify previously unknown dendritic cell and monocyte subsets in human blood, which were indistinguishable in bulk RNA-seq data4. |
| Sample Input Requirement |
Higher |
Lower |
Picelli et al. (2014) developed Smart-seq2, a single-cell RNA-seq method capable of generating full-length cDNA from as little as 10 pg of total RNA, far less than typical bulk RNA-seq requirements5. |
| Rare Cell Type Detection |
Limited |
Possible |
Grün et al. (2015) used single-cell RNA-seq to identify rare enteroendocrine cell types in the mouse intestine, which were masked in bulk sequencing due to their low abundance6. |
| Gene Detection Sensitivity |
Higher |
Lower |
Chen et al. (2019) found that bulk RNA-seq detected more genes per sample (median 13,378) compared to single-cell RNA-seq (median 3,361) in matched human peripheral blood mononuclear cell samples7. |
| Splicing Analysis |
More comprehensive |
Limited |
Ntranos et al. (2019) developed DISCO-seq, combining bulk and single-cell RNA-seq to improve detection of alternative splicing events in complex tissues. |
Other differences between Bulk RNA-seq and scRNA-seq (Farhan Chaudhry et al,.2019)
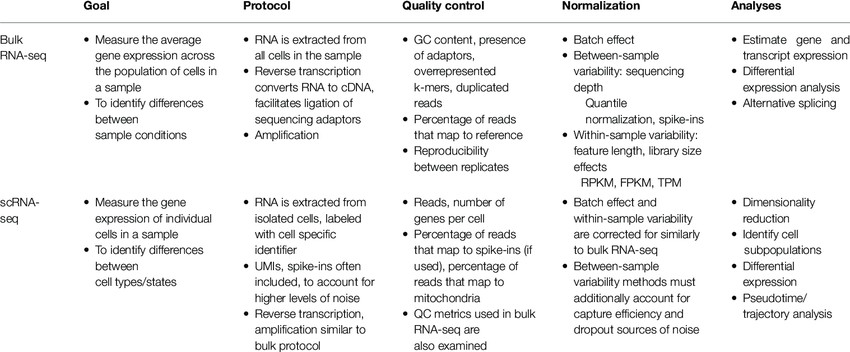
The major differences lie in the resolution, cost, and data complexity. Bulk sequencing provides an average profile, which is useful for understanding the overall gene expression of a tissue, while single-cell sequencing can uncover the nuanced differences between individual cells.
For Differences among scRNA-seq, bulk RNA sequencing and spatial transcriptome sequencing, check out this article scRNA-seq vs. Bulk RNA-seq vs. Spatial Transcriptomics.
4. Applications and Use Cases
Each sequencing method is suitable for different research needs, as evidenced by numerous scientific studies.
Bulk RNA Sequencing
Bulk RNA sequencing is best for large-scale studies and when studying tissues with a homogeneous population of cells.
Cancer research:
Bulk RNA-Seq has proven invaluable in cancer studies. A comprehensive analysis of nearly 7,000 cancer samples from The Cancer Genome Atlas used bulk RNA-Seq to detect novel and clinically relevant gene fusions involved in cancer. This large-scale transcriptome analysis led to the discovery of numerous novel and recurrent kinase gene fusions, many of which now have approved or exploratory drugs available.
Gene expression studies:
Bulk RNA-Seq is widely used for examining the complete set of RNA transcripts produced by a genome. It can reveal whether mutations result in transcriptomic changes that either drive cancer or act as passenger mutations. This approach has been crucial in differentiating cancer subtypes, assessing the impact of mutations, and identifying biomarkers.
Biomarker discovery:
A study on pancreatic cancer used both RNA sequencing and small RNA sequencing to identify differential expression of simple repetitive sequences (SSRs). The researchers found that the frequency of SSR motifs changed dramatically, suggesting potential as tumor biomarkers.
Single-Cell RNA Sequencing
Single-cell RNA sequencing is ideal for studying complex tissues, identifying rare cell types, and exploring the heterogeneity within a population of cells.
Rare cell type identification:
In a study of mouse embryonic stem cells, single-cell RNA-Seq identified a cluster of 3 cells that highly expressed Zscan4 genes. This finding revealed a rare subpopulation of mouse ESCs with greater differentiation potential than previously thought.
Immune profiling:
Single-cell RNA-Seq has been instrumental in identifying new subpopulations of immune cells. For example, it has led to the discovery of new innate lymphoid cell subsets and dendritic cell and monocyte subsets. This level of detail is crucial for understanding how different immune cells respond to disease and for developing targeted therapies.
Cancer heterogeneity:
A recent single-cell RNA-seq study of 49 samples of metastatic lung cancer revealed changes in plasticity induced by non-small cell lung cancer. This type of analysis provides insights into tumor heterogeneity that are not possible with bulk sequencing.
Rare disease-associated cells:
Single-cell RNA-Seq identified CFTR-expressing pulmonary ionocytes, which occur at a rate of 1 in 200 human lung epithelial cells, as potential mediators of cystic fibrosis pathology. Even rarer cell types have been uncovered, such as a population of CAR T cells accounting for only ~1 in 10,000 cells in an infusion product, which contained high human herpesvirus 6 transcriptional activity.
For more in-depth applications, visit our Single-Cell RNA Sequencing: Introduction, Methods, and Applications page.
5. Cost and Efficiency Comparison
Cost is one of the most important factors in deciding between bulk and single-cell sequencing.
- Bulk RNA Sequencing: The cost is much lower, typically about 1/10th of single-cell RNA sequencing. It is the preferred method when researchers need to analyze large numbers of samples at a relatively low cost.
- Single-Cell RNA Sequencing: Due to its complexity and the need to isolate individual cells, it comes at a higher cost. However, its ability to detect rare cell types and cellular heterogeneity makes it an invaluable tool for cutting-edge research.
As both technologies evolve, prices for single-cell sequencing are gradually decreasing, making it more accessible for a wider range of applications.
6. Challenges and Limitations
Challenges and LimitationsBoth bulk RNA sequencing and single-cell RNA sequencing methods come with their own set of challenges:
Bulk RNA Sequencing:
- Limited information on cell heterogeneity: Bulk RNA sequencing misses out on rare or subpopulations of cells that may be crucial to research. For example, a study by Patel et al. demonstrated that bulk RNA sequencing of glioblastoma samples failed to capture the intratumoral heterogeneity that was revealed by single-cell RNA sequencing, potentially missing important insights into tumor progression and treatment resistance.
- Not suitable for complex tissues: If a sample is highly heterogeneous, bulk RNA sequencing may mask important details. This was highlighted in a study by Baron et al., where bulk RNA sequencing of pancreatic tissue failed to identify rare cell types and subtle differences between cell states that were crucial for understanding pancreatic development and disease.
Single-Cell RNA Sequencing:
- Data complexity: Single-cell data is more complicated to process and analyze, requiring sophisticated computational methods. Lähnemann et al. reviewed the challenges in single-cell RNA sequencing data analysis, emphasizing the need for specialized tools to handle the high dimensionality and sparsity of the data.
- Technical challenges: Handling dropout events, where some cells' RNA isn't captured correctly, and isolating individual cells with high fidelity can be difficult. A study by Kharchenko et al. addressed the issue of dropout events in single-cell RNA sequencing data, proposing computational methods to mitigate their effects on downstream analyses.
- Higher cost: The technology and methods used for single-cell RNA sequencing make it more expensive. Stark et al. reported that the cost per sample for single-cell RNA sequencing can be up to ten times higher than bulk RNA sequencing, which can limit its widespread adoption, especially in large-scale studies.
7. Future Trends and Advancements
The future of sequencing technologies looks promising for both bulk and single-cell sequencing. Here are some trends to watch:
- Cost Reduction: Both technologies are expected to become more affordable as advancements in technology reduce the costs associated with sequencing and data analysis.
- Improved Data Integration: Multi-omics approaches that combine single-cell RNA-Seq with other techniques like scATAC-Seq (chromatin accessibility) or scCITE-Seq (protein profiling) are gaining traction, providing a more comprehensive view of cellular functions.
- Hybrid Approaches: Combining bulk and single-cell sequencing can provide both a broad overview and detailed insights into complex biological systems.
For more about future advancements in RNA sequencing technologies, check out this article on RNA Sequencing Technologies.
8. Choosing the Right Approach for Your Research
When deciding between bulk RNA sequencing and single-cell RNA sequencing, consider the following:
Research Goal:
If you need to analyze large, homogeneous populations of cells or tissues, bulk RNA sequencing is the most cost-effective solution.
If you're studying a complex tissue, identifying rare cell types, or exploring cellular heterogeneity, single-cell RNA sequencing will provide more detailed and insightful results.
Budget:
Bulk RNA sequencing is more affordable, making it ideal for large-scale studies or when budget constraints are a factor.
Data Complexity:
If you're prepared to deal with complex datasets and advanced data analysis, single-cell RNA sequencing will offer the resolution and insights needed.
Sample Input:
Consider the amount of starting material available. Single-cell RNA-seq methods like Smart-seq2 can generate full-length cDNA from as little as 10 pg of total RNA, far less than typical bulk RNA-seq requirements.
Cellular Resolution:
If understanding cell-to-cell variability is crucial for your research, single-cell sequencing is the better choice. It can reveal intratumoral heterogeneity and identify clinically relevant subpopulations, which is not possible with bulk sequencing.
Time and Resources:
Analyzing bulk RNA-seq data is generally faster and easier than single-cell data. One researcher mentioned that bulk RNA-seq analysis might take an afternoon, while single-cell analysis could take a week.
Validation:
Consider using both methods complementarily. You might start with bulk sequencing to identify potential targets or markers, and then use single-cell sequencing to validate and explore these findings at a higher resolution.
Need help choosing the right approach? CD Genomics offers expert guidance to help you select the best sequencing strategy for your research. Contact us to learn more about our RNA Sequencing and Single-Cell Sequencing services.
Conclusion: Make the Right Choice with CD Genomics
In summary, bulk RNA sequencing and single-cell RNA sequencing each have their own strengths and are suited for different types of research. Bulk RNA sequencing is ideal for large-scale studies with homogeneous samples, while single-cell sequencing offers unparalleled resolution and the ability to explore cellular heterogeneity.
At CD Genomics, we provide both sequencing services, allowing you to select the most appropriate method based on your specific needs. Our experienced team can help you maximize the value of your sequencing data with comprehensive bioinformatics analysis.
References:
- Wang Y, Mashock M, Tong Z, Mu X, Chen H, Zhou X, Zhang H, Zhao G, Liu B, Li X. Changing Technologies of RNA Sequencing and Their Applications in Clinical Oncology. Front Oncol. 2020 Apr 9;10:447. doi: 10.3389/fonc.2020.00447.
-
Hong, M., Tao, S., Zhang, L. et al. RNA sequencing: new technologies and applications in cancer research. J Hematol Oncol 13, 166 (2020). https://doi.org/10.1186/s13045-020-01005-x
-
Jiang, L., Chen, H., Pinello, L., & Yuan, G.-C. (2016). GiniClust: Detecting rare cell types from single-cell gene expression data with Gini index. Genome Biology, 17, Article 144. https://doi.org/10.1186/s13059-016-1010-4
-
Nguyen, A., Khoo, W. H., Moran, I., Croucher, P. I., & Phan, T. G. (2018). Single cell RNA sequencing of rare immune cell populations. Frontiers in Immunology, 9. https://doi.org/10.3389/fimmu.2018.01553
-
Patel, A. P., Tirosh, I., Trombetta, J. J., Shalek, A. K., Gillespie, S. M., Wakimoto, H., Cahill, D. P., Nahed, B. V., Curry, W. T., Martuza, R. L., Louis, D. N., Rozenblatt-Rosen, O., Suvà, M. L., Regev, A., & Bernstein, B. E. (2014). Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science, 344(6190), 1396-1401. https://doi.org/10.1126/science.1254257
-
Baron, M., Veres, A., Wolock, S. L., Faust, A. L., Gaujoux, R., Vetere, A., Ryu, J. H., Wagner, B. K., Shen-Orr, S. S., Klein, A. M., Melton, D. A., & Yanai, I. (2016). A Single-Cell Transcriptomic Map of the Human and Mouse Pancreas Reveals Inter- and Intra-cell Population Structure. Cell Systems, 3(4), 346-360.e4. https://doi.org/10.1016/j.cels.2016.08.011
-
Lähnemann, D., Köster, J., Szczurek, E., McCarthy, D. J., Hicks, S. C., Robinson, M. D., Vallejos, C. A., Campbell, K. R., Beerenwinkel, N., Mahfouz, A., Pinello, L., Skums, P., Stamatakis, A., Attolini, C. S.-O., Aparicio, S., Baaijens, J., Balvert, M., Barbanson, B. de, Cappuccio, A., … Schönhuth, A. (2020). Eleven grand challenges in single-cell data science. Genome Biology, 21(1), 31. https://doi.org/10.1186/s13059-020-1926-6
-
Kharchenko, P. V., Silberstein, L., & Scadden, D. T. (2014). Bayesian approach to single-cell differential expression analysis. Nature Methods, 11(7), 740-742. https://doi.org/10.1038/nmeth.2967
-
Stark, R., Grzelak, M., & Hadfield, J. (2019). RNA sequencing: the teenage years. Nature Reviews Genetics, 20(11), 631-656. https://doi.org/10.1038/s41576-019-0150-2
-
Patel, A. P., Tirosh, I., Trombetta, J. J., Shalek, A. K., Gillespie, S. M., Wakimoto, H., Cahill, D. P., Nahed, B. V., Curry, W. T., Martuza, R. L., Louis, D. N., Rozenblatt-Rosen, O., Suvà, M. L., Regev, A., & Bernstein, B. E. (2014). Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science, 344(6190), 1396-1401. https://doi.org/10.1126/science.1254257
-
Ziegenhain, C., Vieth, B., Parekh, S., Reinius, B., Guillaumet-Adkins, A., Smets, M., Leonhardt, H., Heyn, H., Hellmann, I., & Enard, W. (2017). Comparative Analysis of Single-Cell RNA Sequencing Methods. Molecular Cell, 65(4), 631-643.e4. https://doi.org/10.1016/j.molcel.2017.01.023
-
Villani, A. C., Satija, R., Reynolds, G., Sarkizova, S., Shekhar, K., Fletcher, J., Griesbeck, M., Butler, A., Zheng, S., Lazo, S., Jardine, L., Dixon, D., Stephenson, E., Nilsson, E., Grundberg, I., McDonald, D., Filby, A., Li, W., De Jager, P. L., ... Hacohen, N. (2017). Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science, 356(6335), eaah4573. https://doi.org/10.1126/science.aah4573
-
Picelli, S., Faridani, O. R., Björklund, Å. K., Winberg, G., Sagasser, S., & Sandberg, R. (2014). Full-length RNA-seq from single cells using Smart-seq2. Nature Protocols, 9(1), 171-181. https://doi.org/10.1038/nprot.2014.006
-
Grün, D., Lyubimova, A., Kester, L., Wiebrands, K., Basak, O., Sasaki, N., Clevers, H., & van Oudenaarden, A. (2015). Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature, 525(7568), 251-255. https://doi.org/10.1038/nature14966
-
Stark, R., Grzelak, M., & Hadfield, J. (2019). RNA sequencing: the teenage years. Nature Reviews Genetics, 20(11), 631-656. https://doi.org/10.1038/s41576-019-0150-2
-
Lähnemann, D., Köster, J., Szczurek, E., McCarthy, D. J., Hicks, S. C., Robinson, M. D., Vallejos, C. A., Campbell, K. R., Beerenwinkel, N., Mahfouz, A., Pinello, L., Skums, P., Stamatakis, A., Attolini, C. S.-O., Aparicio, S., Baaijens, J., Balvert, M., Barbanson, B. de, Cappuccio, A., ... Schönhuth, A. (2020). Eleven grand challenges in single-cell data science. Genome Biology, 21(1), 31. https://doi.org/10.1186/s13059-020-1926-6


 Sample Submission Guidelines
Sample Submission Guidelines
 Figure 1. Applications of single-cell and bulk RNA sequencing in onco-immunology(Maria Kuksin et al,. 2021)
Figure 1. Applications of single-cell and bulk RNA sequencing in onco-immunology(Maria Kuksin et al,. 2021) Figure 2. The single cell RNASeq. (Ye Wang et al,.2020)
Figure 2. The single cell RNASeq. (Ye Wang et al,.2020)
