T-Cell Receptor (TCR) sequencing has emerged as a transformative methodology in the comprehensive study of the immune system. By enabling the detailed analysis of T cell diversity and function, TCR sequencing offers critical insights into T cells, which are indispensable components of the immune response. This article delineates the fundamental aspects of TCR sequencing, elucidating its diverse applications and the underlying techniques that facilitate its operation. Such a discourse aims to elucidate the profound impact that TCR sequencing is having on the fields of immunology and personalized medicine, thus highlighting its potential to reshape our understanding and treatment of various diseases.
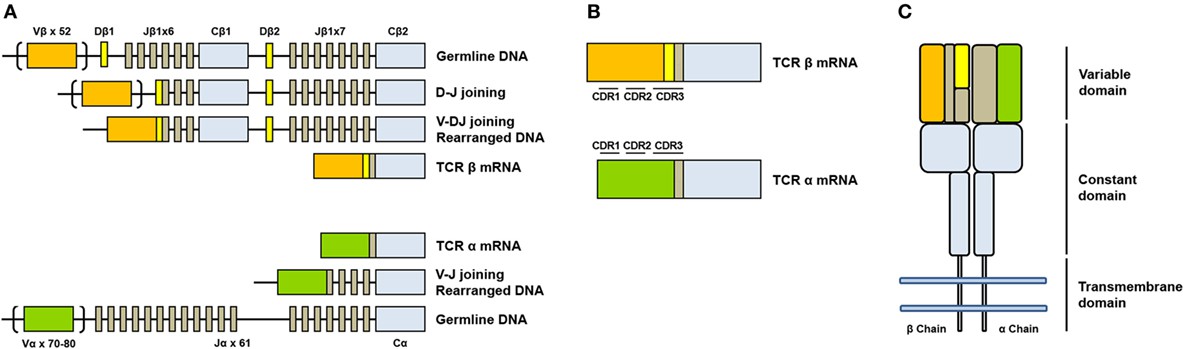 Figure 1. Somatic V(D)J arrangement in the alpha and beta chains. (De Simone et al,.2018)
Figure 1. Somatic V(D)J arrangement in the alpha and beta chains. (De Simone et al,.2018)
Key Table: Why TCR Sequencing Matters
| Aspect |
Details |
| What it is |
A method to sequence the T cell receptor (TCR) to track immune responses. |
| Key Applications |
Cancer immunotherapy, infectious disease research, autoimmune disease studies, transplantation monitoring. |
| Techniques |
Bulk sequencing and single-cell sequencing. |
| Market Growth |
Significant global growth in TCR sequencing technology, especially in cancer immunotherapy. |
| Cost Considerations |
Bulk sequencing is cost-effective; single-cell sequencing provides deeper insights but at a higher cost. |
What is TCR Sequencing?
TCR sequencing is a molecular technique designed to identify and track individual T cells and their clonal populations. This method primarily targets the complementarity-determining region 3 (CDR3) of the TCR, a region characterized by considerable variability and critical for antigen recognition. Typically, both the α (alpha) and β (beta) chains of the TCR are analyzed, as they jointly contribute to the receptor's ability to recognize a wide array of antigens. Given their central role in the adaptive immune response, the sequencing of these chains provides crucial insights into the diversity and functional dynamics of the immune system, thereby enhancing our understanding of immune recognition, tolerance, and response.
Why is TCR Sequencing Important?
TCR sequencing is an important tool that helps us understand how our immune system works to fight off threats like viruses, cancer, and other harmful invaders. It does this by looking at specific parts of T cells, which are a key component of the immune system. By studying T cell clones, researchers can track how the immune system responds to infections, monitor how diseases progress, and find better ways to treat them. This technology is changing how we personalize treatments, making it easier to develop therapies that work for each individual based on their unique immune system.
Key Applications of TCR Sequencing
TCR sequencing has many uses, both in research and in medical practice. Here are some key ways it's being applied:
Cancer Immunotherapy
TCR sequencing is important in cancer treatment, especially when it comes to immunotherapy. This type of therapy helps the immune system fight cancer. TCR sequencing can identify T cells that are reacting to cancer cells. Researchers can then track these T cells to see if they are growing and fighting the tumor. For example, in a study of lung cancer patients, TCR sequencing helped find T cells that were specifically attacking cancer cells, offering clues for improving treatment(ÁF Sanromán et al,. 2023).
 Figure 2. Schematic representation of TCR metrics in immuno-oncology measured by TCR sequencing. (ÁF Sanromán et al,. 2023)
Figure 2. Schematic representation of TCR metrics in immuno-oncology measured by TCR sequencing. (ÁF Sanromán et al,. 2023)
Infectious Disease Research
Our immune system responds differently to different infections, whether it's a virus or bacteria. TCR sequencing allows researchers to track how T cells respond to these infections. This helps scientists understand how well the immune system is working to fight off the infection. In a study on the M. tuberculosis, researchers used TCR sequencing to find specific T cells that were helping to control the infection, providing valuable information for vaccine development(Munyaradzi Musvosvi et al,. 2023).
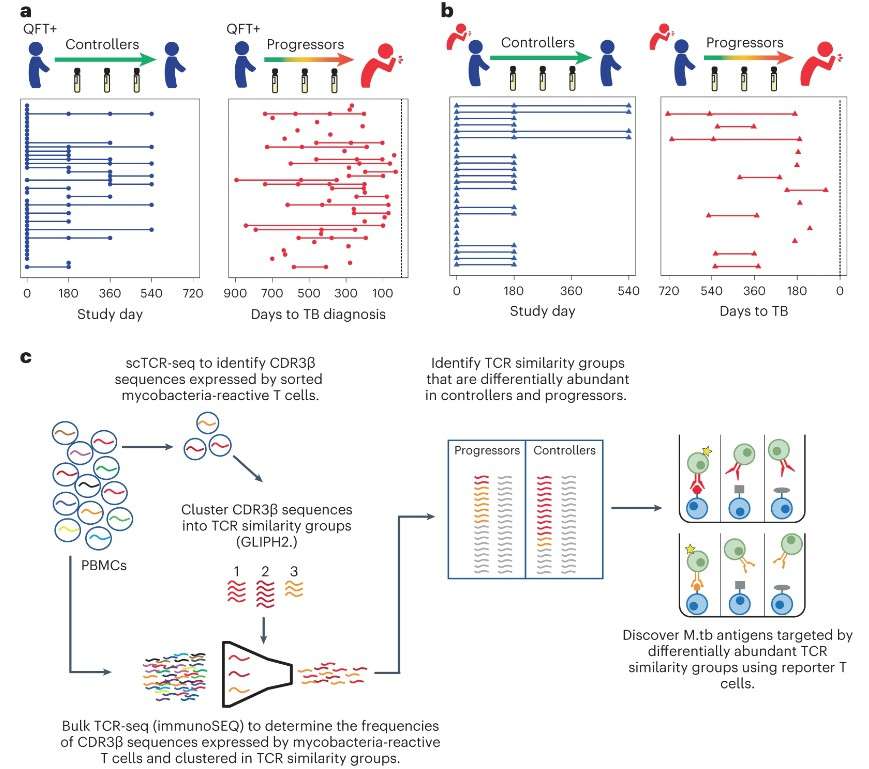 Figure 3. Identification of TCR sequences and antigens recognized by M. tuberculosis lysate-responsive T cells in controllers and progressors.(Munyaradzi Musvosvi et al,. 2023).
Figure 3. Identification of TCR sequences and antigens recognized by M. tuberculosis lysate-responsive T cells in controllers and progressors.(Munyaradzi Musvosvi et al,. 2023).
Autoimmune Disease Diagnostics
In autoimmune diseases, the immune system mistakenly attacks the body's own tissues. TCR sequencing can help identify which T cells are causing the problem. By studying these T cells, scientists can find new ways to treat autoimmune diseases. For instance, in systemic lupus erythematosus (SLE) patients with lupus nephritis, TCR sequencing identified significant differences in TCR diversity and usage of TRBV/TRBJ genes compared to healthy controls(Xiaolan Ye et al,. 2020.
Transplantation Monitoring
After an organ transplant, doctors need to make sure the immune system doesn't attack the new organ. TCR sequencing can help doctors monitor the immune response and detect early signs of organ rejection. For example, by regularly checking the T cells of a heart transplant recipient, doctors could identify if the immune system is starting to attack the new heart, allowing them to intervene before rejection becomes serious.
Understanding Aging Processes
As we get older, our immune system changes and can become less effective. TCR sequencing can help researchers understand how T cells change with age, which could lead to better treatments to keep the immune system healthy in older adults. For example, researchers might study older people's T cells over time to learn more about how aging affects immunity and how to improve health outcomes.
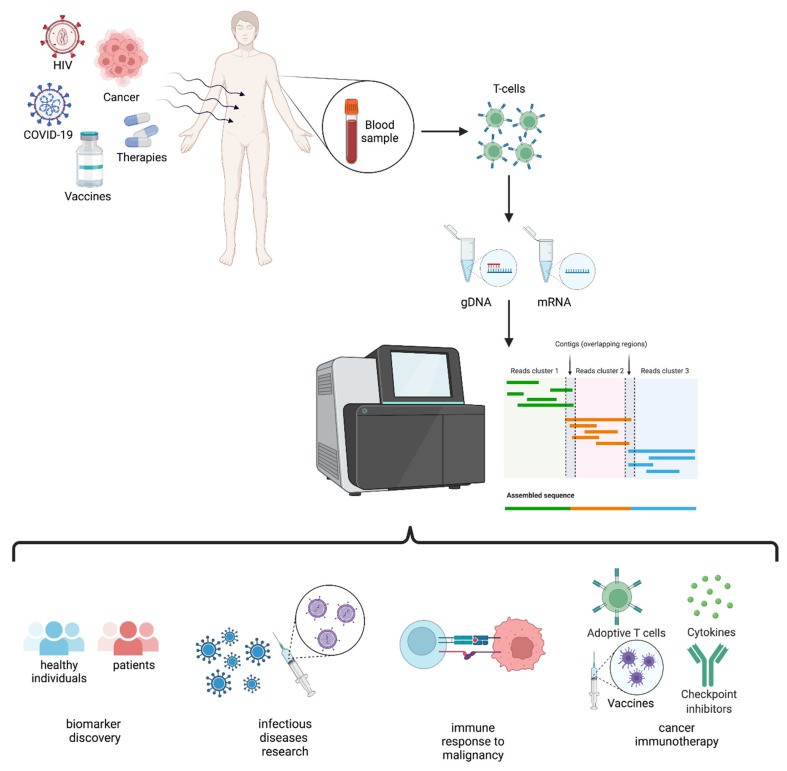 Figure 4. Schematic representation of TCR repertoire analysis and applications. (Lucia Mazzotti et al,. 2022)
Figure 4. Schematic representation of TCR repertoire analysis and applications. (Lucia Mazzotti et al,. 2022)
Comparison of TCR Sequencing Techniques
There are two main approaches to TCR sequencing: bulk sequencing and single-cell sequencing. Each technique offers unique advantages depending on the research goals.
Bulk sequencing analyzes T-cell populations as a whole, providing a broad view of TCR diversity and clonal expansion.
In a study of multiple sclerosis (MS) patients, researchers used bulk TCR sequencing to analyze cerebrospinal fluid samples. They identified expanded T-cell clones associated with disease activity, revealing a correlation between clonal expansion and clinical progression. This approach allowed for the identification of disease-specific TCR signatures across a large cohort of patients.
Single-cell sequencing examines individual T-cells, preserving information about paired α and β chains and allowing for more detailed analysis of specific T-cell subsets.
A single-cell TCR sequencing study of tumor-infiltrating lymphocytes in colorectal cancer patients revealed unexpected diversity in T-cell phenotypes. Researchers identified a subset of CD8+ T-cells with a unique transcriptional profile and TCR sequences associated with improved patient outcomes. This level of resolution would not have been possible with bulk sequencing alone(Frank et al,. 2023).
| Feature |
Bulk Sequencing |
Single-Cell Sequencing |
| Sample Analysis |
T-cell populations |
Individual T-cells |
| Sequence Coverage |
Higher |
Lower |
| αβ-TCR Pairing |
Lost |
Preserved |
| Cost |
Lower |
Higher |
| Ideal For |
Immune repertoire diversity |
TCR specificity studies |
- Bulk Sequencing: Best for analyzing large populations of T cells. It's cost-effective and suitable for studying immune diversity in large groups.
- Single-Cell Sequencing: Provides deeper insights by analyzing individual T cells. It's ideal for studying TCR specificity and understanding the behavior of individual T-cell clones.
Complementary Approaches
Some studies combine both methods for comprehensive analysis. For instance, in a study of SARS-CoV-2-specific T-cells, researchers first used bulk sequencing to identify expanded clones, then employed single-cell sequencing to characterize the transcriptional profiles and paired TCR sequences of these clones. This approach provided both breadth and depth in understanding the T-cell response to viral infection.
Advancements in TCR Sequencing Techniques
Recent breakthroughs in sequencing technologies have made TCR sequencing more accurate, affordable, and accessible. Next-generation sequencing (NGS) platforms have improved sequence accuracy and read depth, making it easier to study even the smallest populations of T cells.
High-Throughput Sequencing:
This technology allows researchers to analyze a large number of samples simultaneously, greatly speeding up the research process.
Multiplex PCR and 5' RACE
Multiplex PCR and 5' Rapid Amplification of cDNA Ends (5' RACE) are two common methods for targeted amplification of TCR sequences.Example: In a study of regulatory CD4+ T cells, Cook et al. (2020) employed 5' RACE PCR to amplify TCRβ chains, followed by Sanger sequencing. This approach allowed for unbiased amplification of TCR genes, avoiding the primer bias associated with multiplex PCR.
RNA-seq for TCR Analysis
RNA-seq provides comprehensive gene expression information alongside immune repertoire data, offering advantages over genomic DNA-based methods.
Researchers studying multiple sclerosis patients used RNA-seq to analyze TCR repertoires in cerebrospinal fluid samples. This approach not only identified expanded T cell clones associated with disease activity but also provided insights into the transcriptional profiles of these cells, revealing potential therapeutic targets.
Single-Cell TCR Sequencing
Single-cell sequencing techniques have revolutionized TCR analysis by preserving information about paired α and β chains.
In a groundbreaking study of colorectal cancer patients, researchers used single-cell TCR sequencing to analyze tumor-infiltrating lymphocytes. They identified a subset of CD8+ T cells with unique TCR sequences and transcriptional profiles associated with improved patient outcomes. This level of detail would not have been possible with bulk sequencing methods.
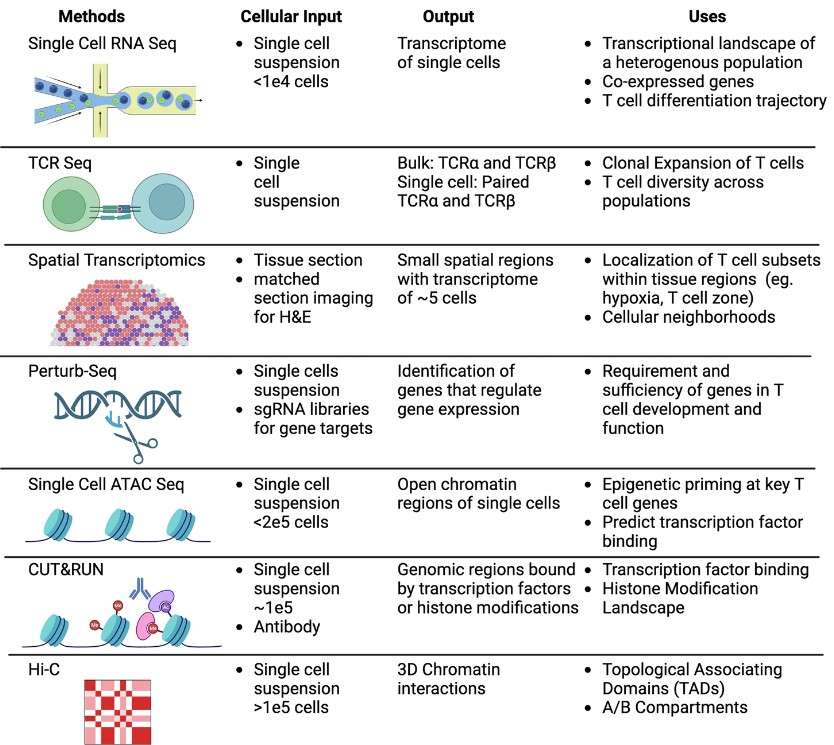 Figure 5.Overview of sequencing methods for investigation of T cell regulation and function. Aaron Yang et al,. 2024)
Figure 5.Overview of sequencing methods for investigation of T cell regulation and function. Aaron Yang et al,. 2024)
Integration of TCR Sequencing with Other Technologies
Combining TCR sequencing with other advanced techniques has opened new avenues for research.
Howie and colleagues developed pairSEQ, a method that accurately pairs TCRα and TCRβ sequences without single-cell isolation. This technique, now available through Adaptive Biotechnologies, allows for high-throughput analysis of TCR diversity in complex samples.
Single-Cell Multi-Omics Approaches
Recent advancements combine TCR sequencing with transcriptomics and protein expression analysis at the single-cell level. The development of CITE-seq and REAP-seq technologies has enabled simultaneous analysis of TCR sequences, transcriptomes, and protein expression in individual T cells. In a study of SARS-CoV-2-specific T cells, researchers used these methods to characterize both the TCR repertoire and the functional states of virus-specific T cells, providing crucial insights into the immune response to COVID-19.These advancements in TCR sequencing techniques have dramatically enhanced our ability to study T cell biology, immune responses, and disease mechanisms at unprecedented resolution.
Enhanced Data Analysis Tools:
Advances in bioinformatics are helping researchers interpret vast amounts of sequencing data, leading to more precise insights. Several innovative tools have emerged to address the challenges of analyzing large-scale TCR sequencing data:
VisTCR:
This client-based HTML program integrates multiple cutting-edge analysis algorithms in a hierarchical fashion. For instance, researchers studying T cell responses in autoimmune diseases could use VisTCR to visualize clonotype distribution across patient samples, revealing expanded T cell clones associated with disease progression.
pyTCR:
This computational notebook-based solution offers comprehensive TCR-Seq data analysis with improved scalability. In a study of COVID-19 patients, researchers might use pyTCR to analyze 46 TCR-Seq samples, comparing T cell repertoires between severe and mild cases to identify potential prognostic markers.
TCRosetta:
This integrated online server combines various analytical methods for TCR repertoire analysis and visualization. For example, cancer immunologists could use TCRosetta to perform large-scale sequence clustering on tumor-infiltrating lymphocytes, identifying shared TCR motifs across patients that may indicate tumor-reactive T cells.
pairSEQ:
This method, developed by Howie and colleagues, accurately pairs TCRα and TCRβ sequences without single-cell isolation. In a hypothetical study of graft-versus-host disease, researchers could use pairSEQ to track the expansion of donor-derived T cell clones in transplant recipients, providing early detection of potential complications.
These advanced tools enable researchers to extract more meaningful insights from TCR sequencing data, facilitating discoveries in immunology and personalized medicine.
Industrial Applications and Market Trends
The global TCR sequencing market is experiencing rapid growth, driven by advances in sequencing technologies and the increasing adoption of immunotherapies in cancer treatment. This market expansion is also fueled by the demand for more precise diagnostic tools and personalized medicine.
- Market Growth: The global TCR sequencing market is projected to grow significantly in the coming years, driven by technological innovations and increasing applications in oncology.
- Emerging Technologies: New advancements, such as next-generation sequencing (NGS) and more affordable sequencing platforms, are making TCR sequencing more accessible and efficient.
Challenges and Future Directions
While TCR sequencing holds tremendous promise, it does come with some challenges:
- Cost: Deep sequencing and single-cell analysis can be expensive, especially for large-scale studies.
- Data Complexity: The vast amount of data generated by TCR sequencing can be overwhelming and requires sophisticated computational tools to interpret.
Looking ahead, advances in sequencing technology, data analysis software, and cost-reduction strategies will help overcome these challenges and make TCR sequencing even more accessible and impactful in medicine.
Conclusion
TCR sequencing represents a cutting-edge approach in the fields of immunology and personalized medicine. TCR sequencing is facilitating new avenues of inquiry and clinical applications across a spectrum of disciplines, from cancer immunotherapy to autoimmune disease studies. As the technology continues to evolve and become more accessible, its potential to transform healthcare is vast.
At CD Genomics, our mission is to provide researchers with high-quality TCR sequencing services, thereby facilitating more informed decision-making. Are you interested in learning how TCR sequencing can facilitate transformative advances in your research? visit our TCR Sequencing Services page.
Frequently Asked Questions (FAQs)
What is the difference between TCR sequencing and BCR sequencing?
TCR sequencing focuses on T cell receptors, which recognize antigens presented by MHC molecules, whereas BCR sequencing analyzes B cell receptors, which bind directly to antigens. Both methods are vital in studying the immune system, but they focus on different aspects of immune recognition.
How deep should TCR sequencing be?
The sequencing depth depends on the sample type and the research goals. For studies involving large, diverse cell populations, deeper sequencing is often necessary. However, this comes with the trade-off of higher costs and lower throughput.
What can TCR sequencing tell us about immune responses?
TCR sequencing can provide valuable insights into clonal expansion, T cell diversity, and antigen-specific responses. It can reveal how T cells respond to infections, cancer, and other diseases, as well as how these responses evolve over time.
How does CD Genomics approach TCR sequencing?
At CD Genomics, we offer comprehensive TCR sequencing services designed to meet the needs of both basic research and clinical applications. Our approach combines cutting-edge sequencing technologies with expert analysis to provide precise, actionable insights into immune function. Learn more about our offerings on our TCR Sequencing page.
References:
- De Simone, M., Rossetti, G., & Pagani, M. (2018). Single-cell T cell receptor sequencing: Techniques and future challenges. Frontiers in Immunology, 9(1638). https://doi.org/10.3389/fimmu.2018.01638
-
Sanromán ÁF, Joshi K, Au L, Chain B, Turajlic S. TCR sequencing: applications in immuno-oncology research. Immunooncol Technol. 2023 Feb 4;17:100373. doi: 10.1016/j.iotech.
-
Ye, X., Wang, Z., Ye, Q., Zhang, J., Huang, P., Song, J., Li, Y., Zhang, H., Song, F., Xuan, Z., & Wang, K. (2020). High-throughput sequencing-based analysis of T cell repertoire in lupus nephritis. Frontiers in Immunology, 11. https://doi.org/10.3389/fimmu.2020.01618
-
Mazzotti L, Gaimari A, Bravaccini S, Maltoni R, Cerchione C, Juan M, Navarro EA, Pasetto A, Nascimento Silva D, Ancarani V, Sambri V, Calabrò L, Martinelli G, Mazza M. T-Cell Receptor Repertoire Sequencing and Its Applications: Focus on Infectious Diseases and Cancer. Int J Mol Sci. 2022 Aug 2;23(15):8590. doi: 10.3390/ijms23158590
-
Rosati, E., Dowds, C.M., Liaskou, E. et al. Overview of methodologies for T-cell receptor repertoire analysis. BMC Biotechnol 17, 61 (2017). https://doi.org/10.1186/s12896-017-0379-9
-
Frank, M. L., Lu, K., Erdogan, C., Han, Y., Hu, J., Wang, T., Heymach, J. V., Zhang, J., & Reuben, A. (2023). T-cell receptor repertoire sequencing in the era of cancer immunotherapy. Clinical Cancer Research, 29(6), 994–1008. https://doi.org/10.1158/1078-0432.CCR-22-2469
-
Mazzotti L, Gaimari A, Bravaccini S, Maltoni R, Cerchione C, Juan M, Navarro EA, Pasetto A, Nascimento Silva D, Ancarani V, Sambri V, Calabrò L, Martinelli G, Mazza M. T-Cell Receptor Repertoire Sequencing and Its Applications: Focus on Infectious Diseases and Cancer. Int J Mol Sci. 2022 Aug 2;23(15):8590. doi: 10.3390/ijms23158590
-
Shen, Y., Voigt, A., Leng, X., Rodriguez, A. A., & Nguyen, C. Q. (2023). A current and future perspective on T cell receptor repertoire profiling. Frontiers in Genetics, 14. https://doi.org/10.3389/fgene.2023.1159109
-
Yang, A., Poholek, A.C. Systems immunology approaches to study T cells in health and disease. npj Syst Biol Appl 10, 117 (2024). https://doi.org/10.1038/s41540-024-00446-1
-
Ni, Q., Zhang, J., Zheng, Z., Chen, G., Christian, L., Grönholm, J., Yu, H., Zhou, D., Zhuang, Y., Li, Q.-J., & Wan, Y. (2020). VisTCR: An interactive software for T cell repertoire sequencing data analysis. Frontiers in Genetics, 11. https://doi.org/10.3389/fgene.2020.00771
-
Peng, K., Moore, J., Vahed, M., Brito, J., Kao, G., Burkhardt, A. M., Alachkar, H., & Mangul, S. (2022). pyTCR: A comprehensive and scalable solution for TCR-Seq data analysis to facilitate reproducibility and rigor of immunogenomics research. Frontiers in Immunology, 13. https://doi.org/10.3389/fimmu.2022.954078
-
Tao Yue, Si-Yi Chen, Wen-Kang Shen, Zhan-Ye Zhang, Liming Cheng, An-Yuan Guo, TCRosetta: An Integrated Analysis and Annotation Platform for T-cell Receptor Sequences, Genomics, Proteomics & Bioinformatics, Volume 22, Issue 4, August 2024, qzae013, https://doi.org/10.1093/gpbjnl/qzae013


 Sample Submission Guidelines
Sample Submission Guidelines
 Figure 1. Somatic V(D)J arrangement in the alpha and beta chains. (De Simone et al,.2018)
Figure 1. Somatic V(D)J arrangement in the alpha and beta chains. (De Simone et al,.2018) Figure 2. Schematic representation of TCR metrics in immuno-oncology measured by TCR sequencing. (ÁF Sanromán et al,. 2023)
Figure 2. Schematic representation of TCR metrics in immuno-oncology measured by TCR sequencing. (ÁF Sanromán et al,. 2023) Figure 3.
Figure 3.  Figure 4. Schematic representation of TCR repertoire analysis and applications. (Lucia Mazzotti et al,. 2022)
Figure 4. Schematic representation of TCR repertoire analysis and applications. (Lucia Mazzotti et al,. 2022) Figure 5.Overview of sequencing methods for investigation of T cell regulation and function. Aaron Yang et al,. 2024)
Figure 5.Overview of sequencing methods for investigation of T cell regulation and function. Aaron Yang et al,. 2024)