T Cell Receptor (TCR) sequencing is a cutting-edge technique that enables researchers to analyze the diversity and specificity of T cell populations. This method plays a critical role in understanding immune responses in various diseases and developing therapies. In this article, we will dive into the principles, methods, technical considerations, and applications of TCR sequencing, shedding light on why it's an essential tool in immunology and clinical research.
Methods of TCR Sequencing
Before delving into TCR sequencing, it's essential to understand the structure of TCRs. For more details, please refer to the article "Differences Between BCR and TCR ".
TCR sequencing can be performed using several methods, each with its own advantages and challenges. The two primary methods are Multiplex PCR (mPCR) and 5' Rapid Amplification of cDNA Ends (5'RACE).
mPCR:
Starting Materials: mPCR can be performed using both DNA or RNA as the starting material. For example, in a study by Robins et al., researchers used genomic DNA from peripheral blood mononuclear cells to analyze the TCR β-chain repertoire in healthy individuals.
PCR Rounds: This method typically involves two PCR rounds, which can amplify specific TCR sequences from a pool of DNA or RNA. A study by Wang et al. demonstrated the use of a two-round PCR approach to amplify and sequence TCR β-chain CDR3 regions from human T cells.
Bias Risk: mPCR is associated with a higher risk of bias in amplification, especially when working with complex samples. Carlson et al. showed that using a large pool of primers in mPCR can lead to amplification biases, particularly for rare TCR sequences.
5' Rapid Amplification of cDNA Ends (5'RACE):
Starting Materials: This method exclusively uses RNA as the starting material. For instance, Mamedov et al. used total RNA extracted from peripheral blood mononuclear cells to perform 5'RACE-based TCR sequencing.
PCR Rounds: Only one PCR round is required, making it a faster and more efficient technique. In a study by Bolotin et al., researchers used a single round of PCR amplification following 5'RACE to analyze the TCR repertoire in human T cells.
Bias Risk: 5'RACE offers a lower risk of bias and is considered more accurate for amplifying TCR sequences from diverse populations. Heather et al. demonstrated that 5'RACE-based methods showed less bias in V gene usage compared to multiplex PCR approaches.
Targeted in-solution enrichment
In addition to Multiplex PCR and 5'RACE, there is another method for TCR sequencing called targeted in-solution enrichment. This method offers some unique advantages and characteristics:
Targeted In-Solution Enrichment
Starting Material: Can use both DNA and RNA
Methodology: Employs RNA baits to capture TCR sequences directly from DNA or RNA sequencing libraries
Process: Captured libraries are re-amplified after the enrichment step
Advantages:
Less biased than multiplex PCR
Can be applied to both DNA and RNA samples, offering flexibility
Allows for capture of full-length TCR sequences
The targeted in-solution enrichment method provides an alternative approach that combines some of the benefits of both mPCR and 5'RACE while mitigating some of their limitations.
Method Comparison Table:
| Feature |
Multiplex PCR |
5'RACE |
Targeted Enrichment |
| Starting Material |
DNA or RNA |
RNA only |
DNA or RNA |
| PCR Rounds |
Typically two |
One |
Variable |
| Bias Risk |
Higher |
Lower |
Intermediate |
| Speed |
Slower |
Faster |
Intermediate |
| Accuracy |
Good |
Better |
Good |
| Full-Length Capture |
Limited |
Yes |
Yes |
This table summarizes the key differences between the two main TCR sequencing methods. Researchers can choose the best approach based on their sample types and the level of precision required for their analysis.
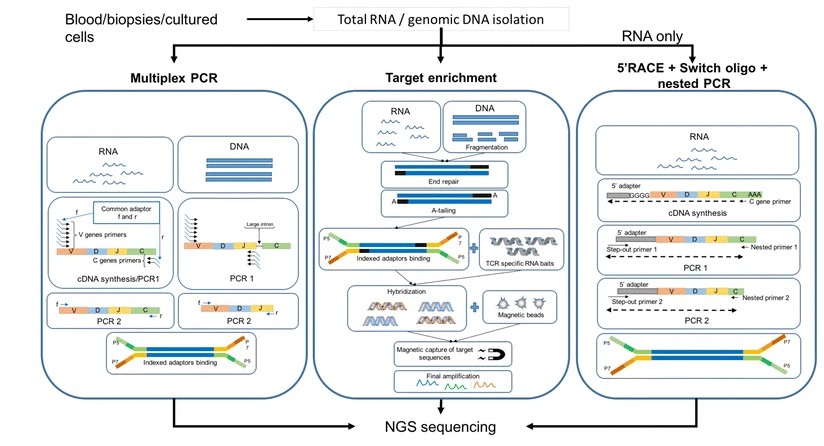 Exemplary workflow of three principal methodologies for TCR library preparation. (Elisa Rosati et al., 2017)
Exemplary workflow of three principal methodologies for TCR library preparation. (Elisa Rosati et al., 2017)
Technical Considerations for TCR Sequencing
Several technical factors must be considered when conducting TCR sequencing, including sample preparation, sequencing depth, and the bioinformatics tools used for analysis.
Sample Preparation:
The type of sample used (e.g., RNA, DNA, or T cells) plays a critical role in the quality and accuracy of TCR sequencing. High-quality RNA or DNA is essential for minimizing errors in amplification and ensuring accurate representation of the TCR repertoire. For instance, a study by Thermo Fisher Scientific emphasizes that obtaining pure and high-quality RNA is crucial for successful RNA-seq sample preparation, as RNA is more unstable than DNA and requires careful handling to prevent degradation (Thermo Fisher Scientific, 2023). Additionally, Mamedov et al. (2013) highlighted the importance of using high-quality RNA extracted from peripheral blood mononuclear cells (PBMCs) for reliable TCR repertoire analysis using 5'RACE methods.
For more on immune repertoire sequencing and the optimal sample types, you can visit our Immune Repertoire Sequencing Introduction.
Sequencing Depth:
Sequencing depth refers to the number of times each sequence is read. Greater sequencing depth increases the likelihood of detecting rare clones, improving the accuracy of immune repertoire analysis. For example, Robins et al. (2012) demonstrated that sequencing 1 million T cells could detect clones at a frequency as low as 1 in 10,000 with 90% power, underscoring the importance of adequate sequencing depth in capturing the full diversity of the TCR repertoire. Furthermore, a study by Bolotin et al. (2013) found that deeper sequencing allowed for more accurate estimation of clonotype frequencies and improved detection of low-frequency TCRs in diverse populations.
If you're interested in learning more about sequencing protocols and optimization strategies, take a look at our TCR Sequencing Using RNA-Seq Protocol.
Bioinformatics Tools:
The analysis of TCR sequencing data involves sophisticated computational tools that can identify patterns and help interpret the immune system's responses. Tools like DeepTCR leverage machine learning to analyze TCR sequences and uncover antigen specificity, providing critical insights for research and clinical applications. A comprehensive comparison of bioinformatics tools for TCR repertoire analysis by Piel et al. (2020) assessed various software packages based on their performance in clonotype detection and error correction accuracy. This study highlighted the need for robust bioinformatics approaches to ensure reliable interpretation of complex TCR sequencing data.In conclusion, careful consideration of sample preparation, sequencing depth, and bioinformatics tools is essential for successful TCR sequencing. These factors collectively influence the accuracy and reliability of the data obtained from TCR repertoire analyses.
Learn more about TCR detection methods and their applications in our TCR Detection: Application and Method blog.
Data Analysis and Interpretation in TCR Sequencing
The data generated by TCR sequencing is complex, and accurate interpretation requires powerful computational tools.
Advanced Computational Tools for TCR Analysis
Tools such as DeepTCR can analyze large datasets of TCR sequences using machine learning algorithms. This analysis reveals patterns related to antigen specificity, helping researchers uncover the immune system's response to diseases, infections, or therapies. For example, a study by Sidhom et al. (2022) demonstrated that DeepTCR could predict responses to immunotherapy with high accuracy by analyzing TCR sequences, effectively linking sequence features to clinical outcomes in cancer patients. This highlights the utility of deep learning frameworks in extracting meaningful insights from complex immunogenomic data.Additionally, the development of pyTCR provides a user-friendly platform for researchers with limited computational backgrounds to analyze TCR-Seq data effectively. According to a study published in Frontiers in Immunology, pyTCR integrates various functionalities such as clonality analysis and diversity metrics, making it easier for researchers to perform comprehensive analyses without extensive programming knowledge (Sidhom et al., 2022). This tool was validated using a dataset from COVID-19 patients, showcasing its practical application in real-world scenarios.Furthermore, VisTCR is another innovative tool that facilitates the analysis of TCR sequencing data through an interactive interface. As reported by Six et al. (2020), VisTCR allows users to group samples for customized analyses and visualize results seamlessly, thus enhancing the interpretability of TCR repertoire data. This capability is particularly beneficial given the increasing volume of TCR sequencing data generated by high-throughput technologies.
Bioinformatics Support at CD Genomics:
At CD Genomics, we provide advanced bioinformatics services to ensure the highest quality analysis of your TCR sequencing data. From error correction to mismatch handling, we ensure that your results are accurate and reliable.
For an overview of next-generation sequencing (NGS) strategies for TCR profiling, check out our detailed Overview of Strategies for TCR Profiling.
Why Choose CD Genomics for TCR Sequencing?
Expertise in Immune Repertoire Analysis:
CD Genomics is a leader in immune repertoire sequencing and TCR sequencing. With advanced technology and expertise, we deliver high-quality, reproducible results for all your research needs.
Comprehensive Services:
We offer a complete solution for TCR sequencing, from experimental design to data interpretation. Our team is with you every step of the way to ensure successful results.
For more details on our TCR sequencing services, visit our TCR-Seq page.
Customizable Solutions:
Our services are tailored to meet the specific needs of your project. Whether you're studying autoimmune diseases, immunotherapy, or transplantation, we can design a custom approach that suits your research goals.
Conclusion: Unlocking the Power of TCR Sequencing
TCR sequencing is an essential tool for advancing our understanding of the immune system. With its ability to analyze the diversity and specificity of T cell populations, this technique opens new doors for cancer research, autoimmune disease studies, immunotherapy development, and more.
References:
-
Bolotin, D. A., Mamedov, I. Z., Britanova, O. V., Zvyagin, I. V., Shagin, D., Ustyugova, S. V., Turchaninova, M. A., Lukyanov, S., Lebedev, Y. B., & Chudakov, D. M. (2012). Next generation sequencing for TCR repertoire profiling: Platform-specific features and correction algorithms. European Journal of Immunology, 42(11), 3073-3083. https://doi.org/10.1002/eji.201242517
- Carlson, C. S., Emerson, R. O., Sherwood, A. M., Desmarais, C., Chung, M.-W., Parsons, J. M., Steen, M. S., LaMadrid-Herrmannsfeldt, M. A., Williamson, D. W., Livingston, R. J., Wu, D., Wood, B. L., Rieder, M. J., & Robins, H. (2013). Using synthetic templates to design an unbiased multiplex PCR assay. Nature Communications, 4, 2680. https://doi.org/10.1038/ncomms3680
- Heather, J. M., Ismail, M., Oakes, T., & Chain, B. (2018). High-throughput sequencing of the T-cell receptor repertoire: pitfalls and opportunities. Briefings in Bioinformatics, 19(4), 554-565. https://doi.org/10.1093/bib/bbw138
- Mamedov, I. Z., Britanova, O. V., Zvyagin, I. V., Turchaninova, M. A., Bolotin, D. A., Putintseva, E. V., Lebedev, Y. B., & Chudakov, D. M. (2013). Preparing unbiased T-cell receptor and antibody cDNA libraries for the deep next generation sequencing profiling. Frontiers in Immunology, 4, 456. https://doi.org/10.3389/fimmu.2013.00456
- Robins, H. S., Campregher, P. V., Srivastava, S. K., Wacher, A., Turtle, C. J., Kahsai, O., Riddell, S. R., Warren, E. H., & Carlson, C. S. (2009). Comprehensive assessment of T-cell receptor β-chain diversity in αβ T cells. Blood, 114(19), 4099-4107. https://doi.org/10.1182/blood-2009-04-217604
- Wang, C., Sanders, C. M., Yang, Q., Schroeder, H. W., Wang, E., Babrzadeh, F., Gharizadeh, B., Myers, R. M., Hudson, J. R., Davis, R. W., & Han, J. (2010). High throughput sequencing reveals a complex pattern of dynamic interrelationships among human T cell subsets. Proceedings of the National Academy of Sciences, 107(4), 1518-1523. https://doi.org/10.1073/pnas.0913939107


 Sample Submission Guidelines
Sample Submission Guidelines
 Exemplary workflow of three principal methodologies for TCR library preparation. (Elisa Rosati et al., 2017)
Exemplary workflow of three principal methodologies for TCR library preparation. (Elisa Rosati et al., 2017)