The emergence of Acetylated RNA Immunoprecipitation Sequencing (acRIP-seq) represents a revolutionary method in understanding RNA molecular modifications. This cutting-edge approach enables researchers to carefully study specific chemical alterations, particularly N4-acetylcytidine (ac4C), which profoundly impacts our comprehension of intricate biological mechanisms.
Epitranscriptomic research has long sought precise techniques for deciphering RNA modification landscapes. acRIP-seq stands at the forefront of this scientific endeavor, facilitating groundbreaking insights across multiple disciplines, including oncological research, neurological investigations, and developmental biology.
Introduction to acRIP-seq
acRIP-seq is a state-of-the-art methodology devised to map RNA acetylation comprehensively across the transcriptome, with a principal focus on the ac4C modification. This technique has garnered substantial interest within the domain of epitranscriptomics, a burgeoning field dedicated to understanding chemical modifications of RNA and their implications for gene regulation.
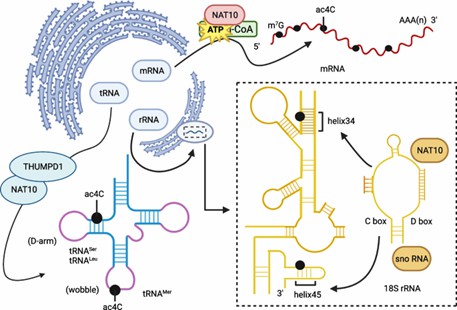 Localization sites of ac4C modification in mRNA, tRNA, and rRNA. (Zhang, S., et al. R2024)
Localization sites of ac4C modification in mRNA, tRNA, and rRNA. (Zhang, S., et al. R2024)
acRIP-seq provides high-resolution mapping of RNA acetylation sites. Researchers can now detect rare modifications with high specificity and sensitivity by combining immunoprecipitation with advanced sequencing technologies.
Initial studies predominantly focused on transfer and ribosomal RNA, demonstrating how ac4C modifications enhance molecular stability and translational accuracy. Recent investigations have expanded this understanding, revealing complex regulatory mechanisms within messenger RNA that significantly influence cellular function.
The methodology's profound implications extend beyond basic research. By illuminating the intricate dynamics of RNA acetylation, acRIP-seq provides unprecedented insights into molecular regulatory networks. Researchers can now explore previously inaccessible biological phenomena, particularly in cancer progression and neurological development.
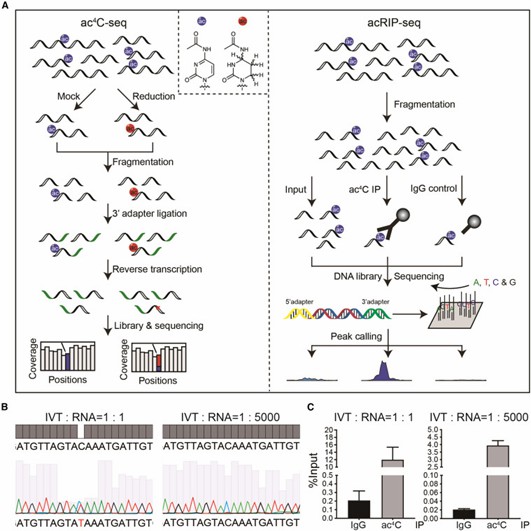 Comparison of two ac4C sequencing methods. (Li, Bin, et al. 2023)
Comparison of two ac4C sequencing methods. (Li, Bin, et al. 2023)
Emerging applications of acRIP-seq show its potential to change our understanding of cellular processes. The technique bridges critical gaps in molecular biology, offering researchers a powerful tool for deciphering complex gene expression mechanisms and potentially developing targeted therapeutic interventions.
As scientific exploration continues, acRIP-seq represents a pivotal technological advancement in epitranscriptomics, promising to unlock deeper comprehension of molecular regulation and cellular complexity.
Principle and Workflow of acRIP-seq
acRIP-seq represents a sophisticated analytical approach for detecting ac4C, a rare yet pivotal RNA modification with profound biological significance. This innovative methodology integrates immunoprecipitation techniques with advanced sequencing strategies, utilizing highly specific antibodies to selectively capture and concentrate RNA fragments containing targeted chemical modifications.
Experimental Workflow and Technical Considerations
Step 1: RNA Fragmentation
The process begins with precise RNA fragmentation, producing segments of about 100 nucleotides. This ensures optimal antibody interaction.
Step 2: Immunoprecipitation
Next, ac4C-specific antibodies bind to the modified RNA fragments, allowing targeted isolation.
Step 3: RNA-antibody Complex Isolation
Magnetic or agarose-based methods are used to separate the RNA-antibody complexes from unmodified RNA.
Step 4: Reverse Transcription
After immunoprecipitation, reverse transcription is used to create complementary DNA (cDNA) for further analysis.
Step 5: Sequencing Library Preparation
The cDNA is then used to prepare sequencing libraries for molecular analysis.
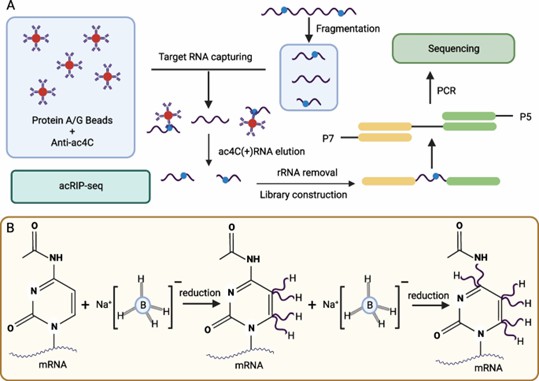 Two methods used to detect ac4C modifications in RNA. A Antibody-based method. B Borohydride reduction method. (Zhang, S., et al. R2024)
Two methods used to detect ac4C modifications in RNA. A Antibody-based method. B Borohydride reduction method. (Zhang, S., et al. R2024)
Analytical Strategies and Computational Approaches
A critical analytical component involves comparative assessment between immunoprecipitated samples and comprehensive input RNA collections. By identifying statistically significant enrichment patterns, researchers can precisely map ac4C modification locations within transcriptional units. These modifications often appear in coding sequences and untranslated regions, offering insights into regulatory mechanisms that affect gene expression and RNA metabolism.
Bioinformatic analysis encompasses multifaceted computational techniques, including:
- Genome-wide sequencing read mapping
- Precise modification site annotation
- Comprehensive regulatory motif identification
Advanced computational methods help researchers study ac4C distribution, biological pathways, and differences across conditions. Such approaches prove particularly valuable in understanding molecular alterations associated with pathological states, including oncological and neurodegenerative contexts.
Broader Implications and Scientific Significance
By delivering both qualitative and quantitative molecular insights, acRIP-seq emerges as a transformative technique for investigating RNA modification landscapes. The methodology provides unprecedented resolution in understanding epitranscriptomic mechanisms, bridging critical knowledge gaps in molecular biology and cellular regulation.
Researchers can now explore intricate RNA modification dynamics with remarkable precision, potentially uncovering novel regulatory mechanisms that govern cellular function and disease progression.
Applications of acRIP-seq in Research
acRIP-seq has emerged as a transformative technology in the exploration of RNA modifications, particularly the study of ac4C and its implications in gene regulation. This methodology has been instrumental in broadening our understanding of RNA biology, offering valuable insights across various domains of research.
RNA Stability and Translational Dynamics
Cutting-edge research utilizing acRIP-seq has unveiled critical insights into ac4C's pivotal role in modulating RNA molecular stability and translational efficiency. Empirical investigations demonstrate that such chemical modifications significantly enhance messenger RNA resilience, thereby optimizing protein synthesis processes. These molecular alterations fundamentally influence cellular functionality, impacting critical biological phenomena including cellular growth, differentiation, and stress response mechanisms.
Pathological Modification Landscapes
The methodology emerges as a sophisticated diagnostic instrument for investigating RNA modification patterns within disease contexts, particularly in oncological and neurodegenerative research domains. By comprehensively mapping transcriptome-wide acetylation modifications, researchers can potentially identify critical biomarkers and therapeutic intervention targets. Specific investigations into ac4C's molecular interactions with oncogenic or tumor suppressor genetic elements provide unprecedented insights into cellular transformation mechanisms.
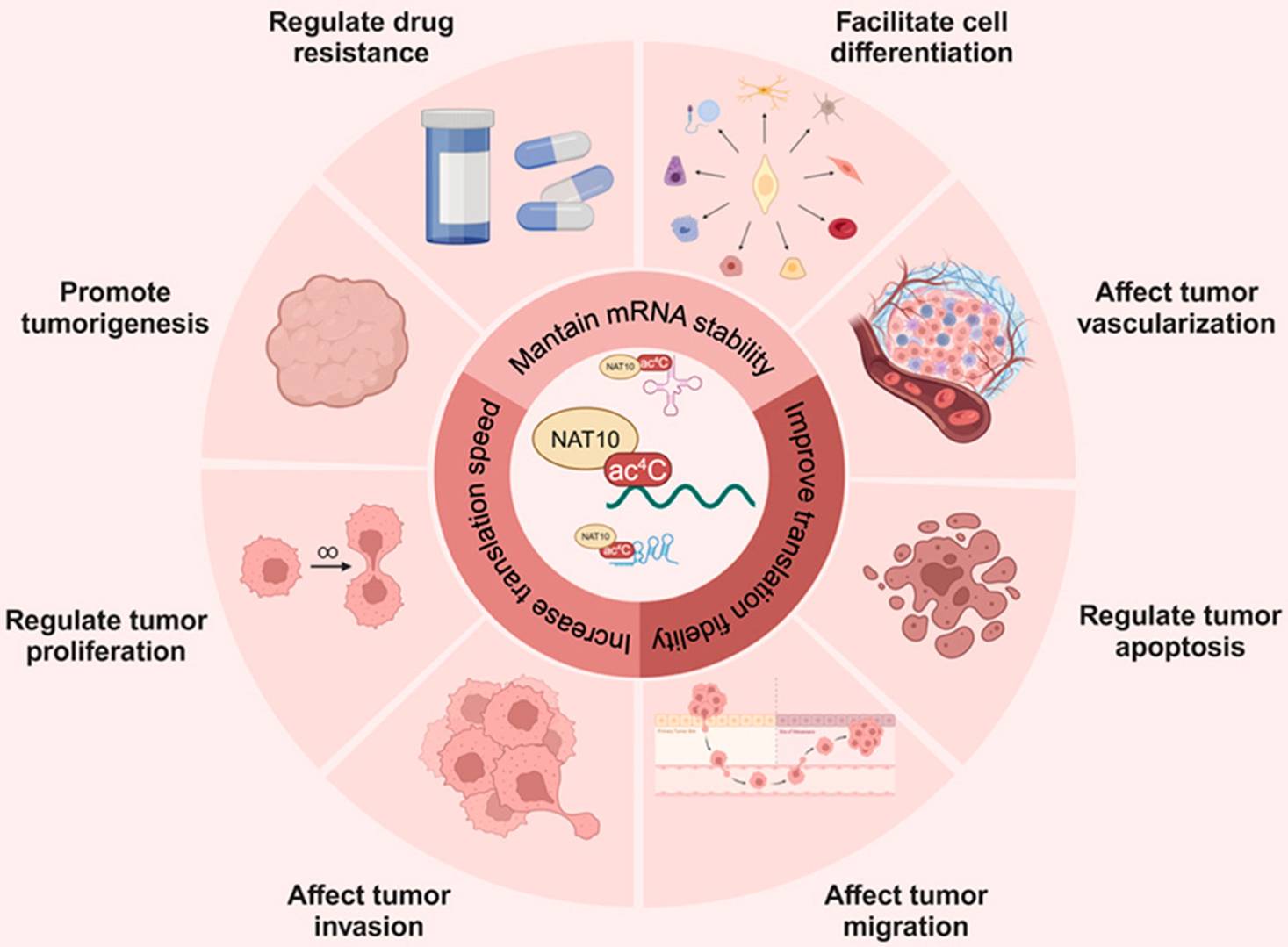 RNA ac4C modification in cancer: Unraveling multifaceted roles and promising therapeutic horizons. (Ouyang, Wenhao, et al. 2024)
RNA ac4C modification in cancer: Unraveling multifaceted roles and promising therapeutic horizons. (Ouyang, Wenhao, et al. 2024)
Expansive Modification Mapping Strategies
acRIP-seq's analytical capabilities extend beyond messenger RNA, encompassing transfer RNA, ribosomal RNA, and diverse non-coding RNA varieties. This comprehensive approach has substantially enriched scientific understanding of RNA modification functional diversity. Researchers have uncovered intricate roles of ac4C in non-coding RNA populations, revealing critical mechanisms in gene regulation, chromatin restructuring, and intercellular communication pathways.
Developmental Biology Investigations
Within developmental biology research, acRIP-seq facilitates nuanced exploration of RNA modification regulatory networks during cellular differentiation and organismal development. Chemical modifications like ac4C profoundly influence mRNA processing, potentially modulating alternative splicing mechanisms and genetic isoform expression. Tracking these molecular interactions enables sophisticated investigations into stem cell differentiation, tissue development, and complex organogenesis processes.
Technological Integration and Multidimensional Analysis
A significant methodological advantage lies in acRIP-seq's capacity for integration with complementary high-throughput technologies. Combining approaches such as RNA sequencing and ribosome profiling allows researchers to establish comprehensive correlations between chemical modifications and gene expression dynamics. This multifaceted analytical strategy provides unprecedented molecular insights into transcript-specific translational mechanisms.
acRIP-seq has emerged as an indispensable investigative platform, revolutionizing our comprehension of RNA modification complexities. By delivering profound transcriptome-wide insights into ac4C's multifaceted roles, this methodology continues to advance molecular biology's frontier, offering researchers powerful tools for deciphering intricate cellular regulatory mechanisms.
Benefits of Using acRIP-seq
acRIP-seq emerges as a groundbreaking technological platform for investigating molecular RNA modifications, particularly N4-acetylcytidine across diverse biological contexts. This comprehensive analytical approach offers multiple distinctive advantages in molecular research:
Precision and Sensitivity in Modification Detection
The methodology distinguishes itself through extraordinary capabilities for identifying RNA modifications present at minimal concentration levels. Utilizing highly specialized antibodies targeting ac4C, researchers can effectively isolate and concentrate modified RNA fragments from intricate biological specimens. This approach ensures the capture of functionally significant molecular variations, even when confronting samples with limited RNA content or complex modification landscapes.
Comprehensive Transcriptome Exploration
Unlike conventional techniques restricted to specific genomic regions, acRIP-seq delivers expansive, genome-wide mapping of chemical modifications. This holistic investigative strategy facilitates comprehensive identification of ac4C across multiple RNA species, including messenger, long non-coding, transfer, and ribosomal RNA varieties. Such extensive profiling substantially enhances our understanding of ac4C's fundamental roles in regulating genetic expression and RNA functionality.
Unbiased Molecular Mapping Strategies
The technique employs an inherently objective approach to modification detection, circumventing requirements for pre-existing locational knowledge of ac4C markers. Leveraging high-affinity immunological tools, researchers can systematically identify modification sites throughout the transcriptome, encompassing both well-characterized and previously undocumented regions. This unrestricted methodology promotes groundbreaking discoveries in RNA molecular biology.
Functional Integration and Multidimensional Analysis
A critical advantage of acRIP-seq lies in its capacity to generate profound functional insights into RNA modifications. By strategically integrating with complementary high-throughput methodologies like RNA sequencing, researchers can establish sophisticated correlations between modification patterns and broader cellular processes. This approach enables comprehensive investigations into translation efficiency, protein synthesis, and genetic regulatory mechanisms.
Optimized Performance with Limited Biological Resources
The technique demonstrates exceptional utility when confronting constrained sample quantities. In specialized research domains such as clinical investigations or rare disease studies, acRIP-seq can efficiently detect ac4C modifications from minimal RNA quantities. This characteristic renders the methodology particularly valuable for examining rare cellular populations, early-stage pathological conditions, and limited patient-derived tissue samples.
Methodological Flexibility and Adaptive Capabilities
acRIP-seq's remarkable adaptability amplifies its research applicability. Investigators can customize the technique to target specific RNA categories or subtypes, ranging from messenger RNA to specialized non-coding RNA varieties. Moreover, the methodology demonstrates remarkable versatility across diverse biological models, spanning cell cultures, tissue specimens, and clinical sample collections.
acRIP-seq represents a transformative analytical platform, synthesizing exceptional sensitivity, comprehensive applicability, and sophisticated data generation capabilities. Its profound potential for delivering intricate molecular insights positions it as an indispensable tool for researchers exploring RNA modification dynamics across complex biological systems.
Recent Advances in acRIP-seq Technology
Recent technological developments in acRIP-seq have dramatically expanded our capacity to investigate RNA modifications, particularly N4-acetylcytidine across diverse molecular landscapes. Breakthrough innovations in antibody design, sequencing platforms, and computational analysis have revolutionized our ability to map chemical modifications with unprecedented precision.
Key Technological Advancements
1. Antibody and Experimental Protocol Refinement: Researchers have developed extraordinarily selective immunological tools targeting ac4C, substantially enhancing detection sensitivity. Sophisticated protocols for RNA fragmentation and molecular binding have been meticulously optimized, minimizing background interference and enabling more reliable transcriptome-wide modification identification.
2. Multidimensional Sequencing Integration: Innovative approaches now seamlessly combine acRIP-seq with complementary sequencing technologies like RNA-seq and ribosome profiling. This integrative methodology provides comprehensive insights into ac4C's multifaceted roles in RNA stability, translational dynamics, and gene expression regulation.
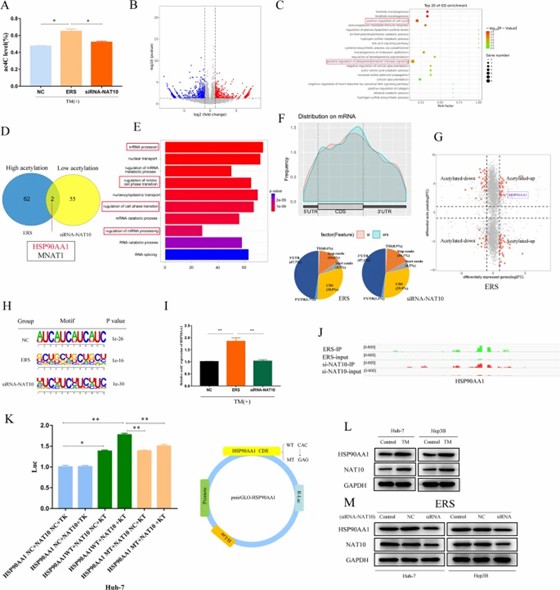 Huh-7 cells induced by TM and knocked down by NAT10 were subjected to liquid chromatography–tandem mass spectrometry, ACRIP-Seq, and RNA-Seq. (Pan, Z., et al. 2023)
Huh-7 cells induced by TM and knocked down by NAT10 were subjected to liquid chromatography–tandem mass spectrometry, ACRIP-Seq, and RNA-Seq. (Pan, Z., et al. 2023)
3. Single-Cell Modification Mapping: Groundbreaking advances in single-cell sequencing technologies have enabled unprecedented resolution in modification analysis. Researchers can now investigate cellular heterogeneity by examining ac4C modifications at individual cellular levels, opening novel investigative pathways in cancer research, neurological studies, and developmental biology.
4. High-Throughput Quantitative Analysis: Advanced sequencing platforms and sophisticated bioinformatics pipelines have transformed acRIP-seq into a highly accessible and quantitative analytical approach. Cutting-edge computational tools facilitate comprehensive assessment of modification abundance, enabling sophisticated peak calling, motif discovery, and cross-dataset integration.
Exemplary Research Investigations
Molecular Translation Dynamics: A seminal investigation published in Cell (2018) explored ac4C's pivotal role in mRNA translational efficiency. Utilizing acRIP-seq and transcriptomic analyses in cellular models, researchers demonstrated how specific cytidine acetylation significantly enhances translation potential and molecular stability.
Oncological Modification Mechanisms: A recent study in Cancer Research (2023) illuminated NAT10's critical function in chemotherapeutic resistance through ac4C-mediated molecular mechanisms. The research uncovered complex interactions between RNA modifications and cellular repair processes, revealing potential therapeutic intervention strategies.
Broader Implications and Future Perspectives
These technological advancements represent a transformative approach to understanding RNA modification landscapes. By providing unprecedented molecular insights, acRIP-seq continues to push the boundaries of our comprehension of cellular regulatory mechanisms, offering promising pathways for both fundamental research and clinical applications.
Statistical Insights and Emerging Industry Trends
The study of RNA modifications, particularly N4-acetylcytidine, is undergoing dynamic expansion, accompanied by significant statistical trends in both academia and industry. These trends underscore the rising significance and diverse applications of acRIP-seq technology across sectors such as academia, biotechnology, and pharmaceutical development.
- Upsurge in Scholarly Publications
Recent years have witnessed a substantial increase in research publications focusing on RNA modifications, with a particular emphasis on ac4C. Investigations into ac4C's roles in gene regulation and its implications in diseases, including cancer and neurodegenerative disorders, are contributing to this surge. These studies underscore the potential of RNA modifications in advancing diagnostic and therapeutic strategies.
- Growing Investment in RNA Sequencing
The global market for RNA sequencing, encompassing acRIP-seq, is expanding at an annual growth rate exceeding 10%. This rapid growth reflects heightened interest from both academic researchers and commercial enterprises in harnessing RNA modifications for drug discovery and biomarker development.
- Rising Demand for Single-Cell Profiling
The application of single-cell RNA sequencing techniques, which include ac4C profiling, is gaining traction. This trend is pivotal in enhancing our understanding of cellular heterogeneity and gene expression dynamics at the level of individual cells, especially in the context of complex diseases.
- Advancements in Bioinformatics Tools
The increasing complexity of acRIP-seq data has spurred a demand for sophisticated bioinformatics tools. These tools are essential for accurately deciphering RNA modification patterns and understanding their functional implications, thereby enhancing the interpretability of vast datasets.
- Integration with Drug Discovery Processes
The biotechnology and pharmaceutical sectors are increasingly leveraging acRIP-seq within drug discovery pipelines. By elucidating RNA modifications such as ac4C, researchers can uncover novel therapeutic targets, advancing the field of precision medicine.
These emerging trends signal the escalating importance of acRIP-seq in both foundational research and industrial innovation, with its transformative potential in unraveling RNA biology and pioneering therapeutic advancement.
Conclusion
acRIP-seq is more than just a sequencing technique; it's a gateway to understanding the intricate world of RNA modifications. Whether you're investigating cancer biology, neuroscience, or developmental biology, acRIP-seq provides the tools you need for groundbreaking discoveries.
At CD Genomics, we're committed to helping researchers achieve their goals with high-quality sequencing services. Reach out to us now and take the next step in your research journey.
References:
- Jiang, L., Chen, H., Pinello, L., & Yuan, G.-C. (2016). GiniClust: Detecting rare cell types from single-cell gene expression data using the Gini index. Genome Biology, 17, article 144. https://doi.org/10.1186/s13059-016-1010-4.
- Arango, D., et al. (2018). Acetylation of Cytidine in mRNA Promotes Translation Efficiency. Cell, 175(7), 1872-1886. https://doi.org/10.1016/j.cell.2018.10.030.
- Xie, R., et al. (2023). NAT10 Drives Cisplatin Chemoresistance by Enhancing ac4C-Associated DNA Repair in Bladder Cancer. Cancer Research, 83(10), 1666-1683. https://doi.org/10.1158/0008-5472.CAN-22-2233.
- Gao, J., Xu, P., Wang, F. et al. Revealing the pharmacological effects of Remodelin against osteosarcoma based on network pharmacology, acRIP-seq and experimental validation. Sci Rep 14, 3577 (2024). https://doi.org/10.1038/s41598-024-54197-4
- Li, Bin, et al. "Transcriptome-wide profiling of RNA N4-cytidine acetylation in Arabidopsis thaliana and Oryza sativa." Molecular Plant 16.6 (2023): 1082-1098. DOI: 10.1016/j.molp.2023.04.009
- Pan, Z., Bao, Y., Hu, M. et al. Role of NAT10-mediated ac4C-modified HSP90AA1 RNA acetylation in ER stress-mediated metastasis and lenvatinib resistance in hepatocellular carcinoma. Cell Death Discov. 9, 56 (2023). https://doi.org/10.1038/s41420-023-01355-8
- Zhang, S., Liu, Y., Ma, X. et al. Recent advances in the potential role of RNA N4-acetylcytidine in cancer progression. Cell Commun Signal 22, 49 (2024). https://doi.org/10.1186/s12964-023-01417-5
- Ouyang, Wenhao, et al. "ac4C RNA modification in Cancer: Unraveling Multifaceted Roles and Promising Therapeutic Horizons." Cancer Letters (2024): 217159. https://doi.org/10.1016/j.canlet.2024.217159


 Sample Submission Guidelines
Sample Submission Guidelines
 Localization sites of ac4C modification in mRNA, tRNA, and rRNA. (Zhang, S., et al. R2024)
Localization sites of ac4C modification in mRNA, tRNA, and rRNA. (Zhang, S., et al. R2024) Comparison of two ac4C sequencing methods. (Li, Bin, et al. 2023)
Comparison of two ac4C sequencing methods. (Li, Bin, et al. 2023) Two methods used to detect ac4C modifications in RNA. A Antibody-based method. B Borohydride reduction method. (Zhang, S., et al. R2024)
Two methods used to detect ac4C modifications in RNA. A Antibody-based method. B Borohydride reduction method. (Zhang, S., et al. R2024) RNA ac4C modification in cancer: Unraveling multifaceted roles and promising therapeutic horizons. (Ouyang, Wenhao, et al. 2024)
RNA ac4C modification in cancer: Unraveling multifaceted roles and promising therapeutic horizons. (Ouyang, Wenhao, et al. 2024) Huh-7 cells induced by TM and knocked down by NAT10 were subjected to liquid chromatography–tandem mass spectrometry, ACRIP-Seq, and RNA-Seq. (Pan, Z., et al. 2023)
Huh-7 cells induced by TM and knocked down by NAT10 were subjected to liquid chromatography–tandem mass spectrometry, ACRIP-Seq, and RNA-Seq. (Pan, Z., et al. 2023)