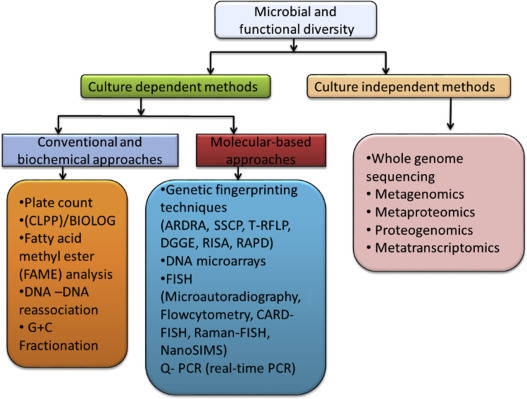
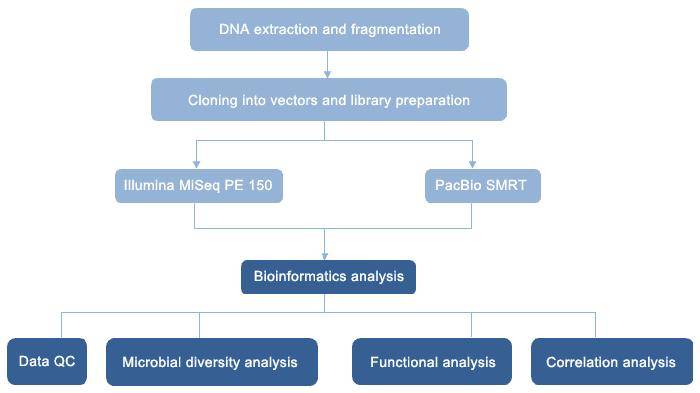
Case Study
Title: How do earthworms affect the pathway of sludge bio-stabilization via vermicomposting
Journal: Science of the Total Environment (IF: 9.8)
Publication Date: January 2024
Sample Type: Municipal Sludge Vermicomposting
Techniques: 16S rDNA sequencing (V3-V4) + 18S rDNA sequencing (V4-V5)
Research: The synergistic interaction between earthworms and microorganisms in vermicomposting systems enhances nitrogen mineralization and organic matter stability. Yet, the specific pathways by which earthworms stabilize the system remain unclear. This study aims to investigate the influence of earthworms on the microbial community and carbon-nitrogen turnover in sludge composting systems. Municipal sludge was composted for 60 days at 20°C, monitoring the trends of organic matter (OM), dissolved organic carbon (DOC), NH4+-N, electrical conductivity (EC), microbial biomass carbon (MBC), and dehydrogenase activity (DHA) over time. Physicochemical evaluation at the end of the treatment revealed significantly lower OM and DOC levels in the vermicomposting group compared to the control group (p < 0.05), with higher EC, NH4+-N, and NO3-N levels (p < 0.05).
Principal component analysis (PCA) proposed potential mechanisms driving carbon and nitrogen turnover by microorganisms during the vermicomposting process. The vermicomposting treatment exhibited higher abundance and diversity of prokaryotic microbial taxa compared to the control, with a more even distribution of microbial community composition, while eukaryotic taxa showed an opposite trend. Earthworm vermicomposting increased microbial abundance involved in organic matter degradation and nitrification, facilitating organic matter transformation and promoting nitrification processes.
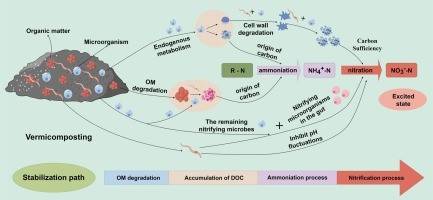 Fig. 2 Potential mechanisms of nitrogen-related transformations during vermicomposting of sewage sludge
Fig. 2 Potential mechanisms of nitrogen-related transformations during vermicomposting of sewage sludge
Title: Changes in soil microbe-mediated carbon, nitrogen and phosphorus cycling during spontaneous succession in abandoned Pb–Zn mining areas
Journal: Science of the Total Environment (IF: 9.8)
Publication Date: April 2024
Sample Type: Soil
Technology: Metagenomics
Research Content: The mechanisms by which soil microbes mediate the cycling of carbon and other nutrients during the restoration process of abandoned mining areas remain unclear. This study utilized soil Metagenomics sequencing to investigate the dynamic changes in soil microbial potential biogeochemical cycling functions during the succession process from Biological soil crust (BSC) to Mixed broad-conifer forest (MBF) in typical abandoned lead-zinc mining areas. Results indicated that soil microbes favored carbon sequestration through anaerobic and microaerobic pathways during succession, with genes controlling carbon degradation and aerobic respiration increasing by 19.56% and 24.79%, respectively, reflecting the dynamic changes in soil microbial carbon cycling genes during succession. In terms of nitrogen cycling, soil microbe-mediated nitrogen cycling genes peaked in the early succession stage, leading to a 75.29% increase in net potential nitrification rate and a 76.81% increase in total nitrogen content. Mantel correlation analysis revealed that TOC, TN, Zn, and Cd content were the primary soil environmental factors influencing soil carbon and phosphorus cycling. Soil AP content emerged as the main influencing factor of genes related to nitrogen cycling. The findings of this study elucidate the dynamic functions of soil microbes during the succession of abandoned Pb-Zn mines, providing insights into biogeochemical cycling mechanisms and underscoring the role of environmental factors in shaping biogeochemical processes in mining area soils.
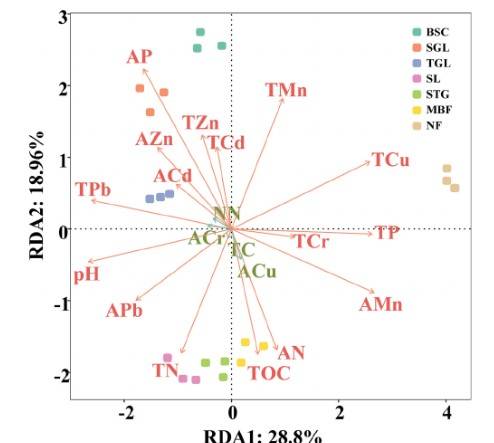 Fig.3 RDA analysis of the correlation between CNP cycling genes and soil factors
Fig.3 RDA analysis of the correlation between CNP cycling genes and soil factors
Title: Farmland Microhabitat Mediated by a Residual Microplastic Film: Microbial Communities and Function
Journal: Environmental Science & Technology (Impact Factor: 11.4)
Publication Date: February 2024
Sample Type: Soil, Microplastic Film
Technology: 16S rDNA Sequencing (V3-V4) + ITS rDNA Sequencing (ITS1) + Metagenomics
Research Content: The impact of residual microplastic films on microhabitats in farmland remains poorly understood. Through 16S rDNA amplicon sequencing, ITS analysis, and metagenomic analysis, this study analyzed the microbial community composition, structure, assembly, and functional genes related to biogeochemical cycling in the plastic film circle and soil at 33 typical farmland sites. Results indicate that residual microplastic films harbor microbial colonization, forming a distinct ecological niche known as the "plastic interface," exhibiting significant differences in microbial community structure and function compared to soil. The residual microplastic film alters the symbiosis and assembly processes of microorganisms. Bacterial community assembly in both the plastic interface and soil is significantly dominated by random processes, while fungal community assembly in the plastic interface is dominantly influenced by random processes, and in soil, it is influenced by deterministic processes. Moreover, the plastic interface formed by residual microplastic films acts as a preferred carrier for pathogens and microbes associated with plastic degradation and nitrogen-sulfur cycling. The abundance of genes related to denitrification and sulfate reduction activity in the plastic circle is notably higher than in soil, thereby increasing the potential risk of nitrogen and sulfur loss. These findings provide valuable insights into understanding the detrimental effects of residual microplastic films on farmland.
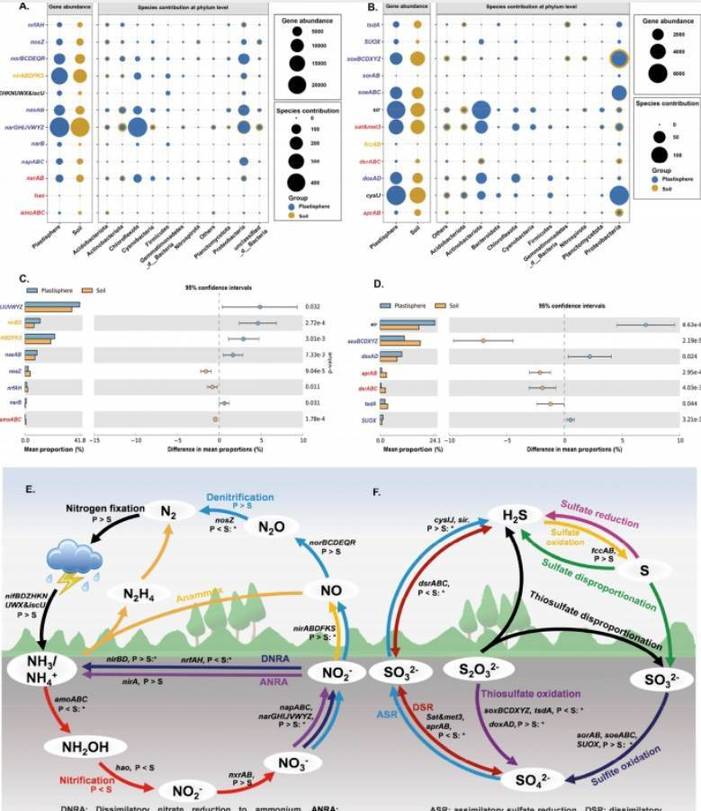 Fig. 4 Gene abundance and microbial (phylum level) contributions to nitrogen and sulfur cycles in soil (n = 12) and plasticsphere (n = 12). (A) Nitrogen cycle, nitrogen fixation (black), nitrification (red), anaerobic ammonium oxidation (orange), denitrification (blue); (B) Sulfur cycle, sulfur oxidation process (blue), assimilatory sulfate reduction process (black), dissimilatory sulfate reduction and oxidation (red), sulfur fixation (orange); (C and D) Relative abundance of genes related to nitrogen cycle (C) and sulfur cycle (D) in soil and plasticsphere based on Wilcoxon rank sum test (p < 0.05); (E and F) Schematic diagram of the effect of residual microplastic film on nitrogen (E) and sulfur (F) cycle processes in farmland (*: p < 0.05)
Fig. 4 Gene abundance and microbial (phylum level) contributions to nitrogen and sulfur cycles in soil (n = 12) and plasticsphere (n = 12). (A) Nitrogen cycle, nitrogen fixation (black), nitrification (red), anaerobic ammonium oxidation (orange), denitrification (blue); (B) Sulfur cycle, sulfur oxidation process (blue), assimilatory sulfate reduction process (black), dissimilatory sulfate reduction and oxidation (red), sulfur fixation (orange); (C and D) Relative abundance of genes related to nitrogen cycle (C) and sulfur cycle (D) in soil and plasticsphere based on Wilcoxon rank sum test (p < 0.05); (E and F) Schematic diagram of the effect of residual microplastic film on nitrogen (E) and sulfur (F) cycle processes in farmland (*: p < 0.05)
Title: Nitrogen and Nod factor signaling determine Lotus japonicus root exudate composition and bacterial assembly
Journal: Nature Communications (IF: 16.6)
Publication Date: April 2024
Sample Types: Soil, Rhizosphere, Roots, Root nodules
Technologies: 16S rDNA sequencing (V5-V7) combined with untargeted metabolomics (root exudates)
Research Content: Symbiosis with nitrogen-fixing bacteria in the soil allows leguminous plants to thrive in nitrogen-deficient environments, impacting the assembly of root microbial communities. However, the influence of interactions between legume hosts and rhizobia on the remaining microbial community, and whether it depends on nitrogen nutrition, remains unclear. This study investigates the role of Nod factor signaling in Lotus japonicus root microbiome assembly through the use of plants and mutants. Utilizing 16S rDNA sequencing and untargeted metabolomics, it was discovered that symbiotic plants produce Nod factors to activate Nod factor signaling in the host, thereby modulating the spectrum of root exudates and the assembly of the symbiotic root microbiome. Lotus japonicus plants with varying symbiotic capabilities grown in unfertilized or nitrate-supplemented soils exhibit three nitrogen-dependent nutritional statuses: starvation, symbiotic, or inorganic. The composition and connectivity of root and rhizosphere microbial communities differ across distinct nitrogen nutritional statuses, indicating varying impacts of symbiosis and inorganic nitrogen on the legume root microbiome. Furthermore, machine learning algorithms were employed to predict bacterial communities with high accuracy in determining the plant's nitrogen nutritional status.
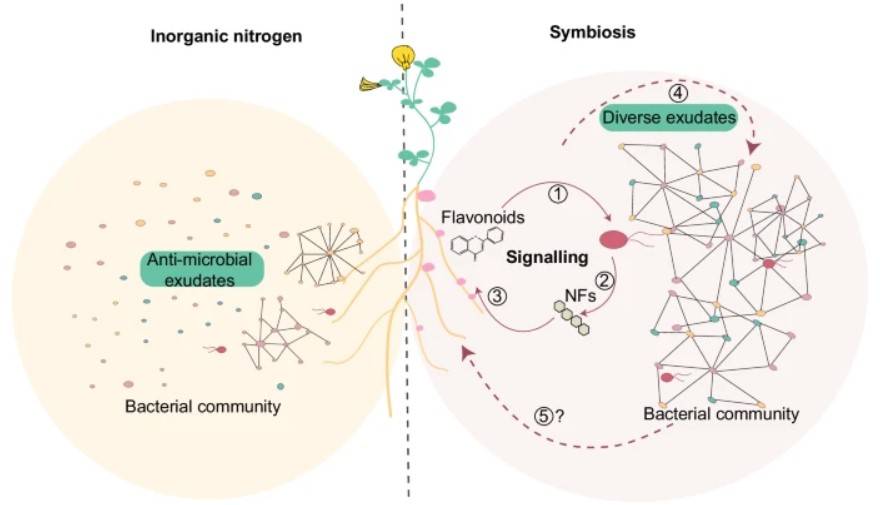 Figure 5. Nitrogen nutrition and signaling during root nodule symbiosis affect community assembly.
Figure 5. Nitrogen nutrition and signaling during root nodule symbiosis affect community assembly.
(Left) Plants grown in the presence of inorganic nitrogen secrete specific metabolites and assemble microbial communities with low connectivity.
(Right) Plants grown under symbiotic permissive conditions secrete metabolites such as flavonoids (1) Metabolites induce nitrogen-fixing rhizobia to produce Nod factors (2) Nod factors are recognized by the host and initiate signaling pathways (3) Adaptation to the symbiont, symbiotic active root secretions change with different conditions (4) Associated microbial communities, and influence on the host to promote symbiotic association and plant growth (5) remain to be determined
Title: Simultaneously enhanced autotrophic–heterotrophic denitrification in iron-based ecological floating bed by plant biomass: Metagenomics insights into microbial communities, functional genes and nitrogen metabolic pathways
Journal: Water Research (Impact Factor: 12.8)
Publication Date: December 2023
Sample Type: Reactor Surface
Technique: Metagenomics
The study aimed to enhance denitrification efficiency in low-pollution water bodies by constructing an ecological floating bed (EFB-IB) loaded with zero-valent iron (ZVI) and plant biomass. Metagenomic sequencing was utilized to analyze the impact of ZVI and plant biomass coupling on microbial community structure, metabolic pathways, and functional genes, elucidating the denitrification mechanism. Results demonstrated that compared to the single ZVI system (EFB-C), EFB-IB significantly improved denitrification efficiency, with average NO3-N removal efficiency increasing by 22.60-59.19% and average NH4+-N removal efficiency increasing by 73.08-91.10%.
Metagenomic analysis revealed the enrichment of microorganisms involved in iron cycling, lignocellulose degradation, and nitrogen metabolism in EFB-IB. The addition of plant biomass simultaneously increased the relative abundance of autotrophic and heterotrophic denitrifying bacteria. Network analysis indicated a synergistic relationship between autotrophic and heterotrophic denitrifying bacteria in EFB-IB. Furthermore, compared to EFB-C, the addition of plant biomass enhanced the relative abundance of genes associated with iron cycling, lignocellulose degradation, and glycolysis processes, ensuring the production of electron donors for both autotrophic and heterotrophic denitrification.
Consequently, EFB-IB exhibited higher relative abundance of key enzymes and functional genes related to denitrification, facilitating NO3-N removal. Additionally, correlation analysis between denitrification and functional genes validated the synergistic mechanism of iron-based autotrophic denitrification and plant biomass-mediated heterotrophic denitrification in EFB-IB. Overall, plant biomass has promising potential for enhancing denitrification efficiency in iron-based EFB systems in low-pollution water bodies.
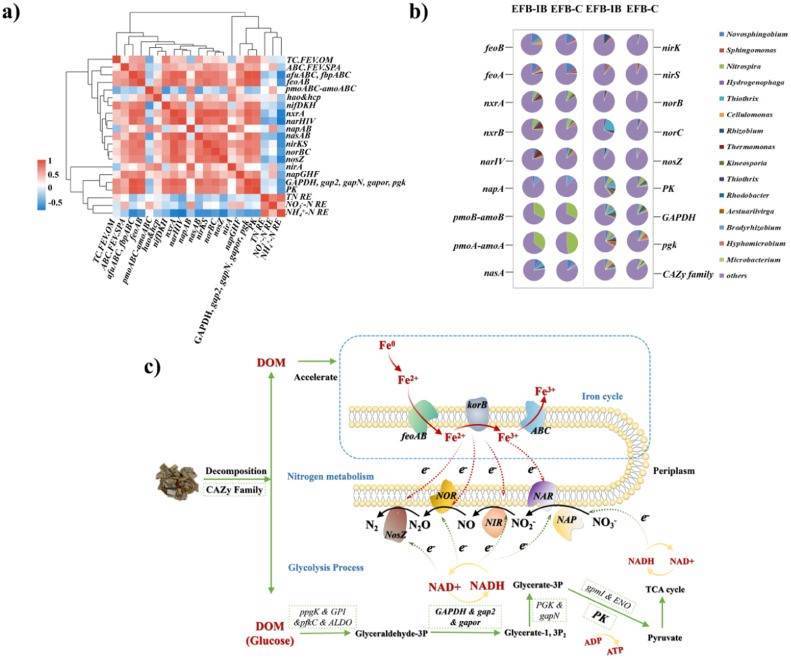 Fig. 6. Spearman correlation analysis between functional genes and denitrification performance (a); Contribution of species to functional genes (b); Possible mechanism of ZVI and plant biomass enhancing denitrification (c)
Fig. 6. Spearman correlation analysis between functional genes and denitrification performance (a); Contribution of species to functional genes (b); Possible mechanism of ZVI and plant biomass enhancing denitrification (c)
References
-
Kuypers M M M, Marchant H K, Kartal B. The microbial nitrogen-cycling network. Nature Reviews Microbiology, 2018, 16(5): 263-276.
- Lei X, Cui G, Sun H, et al. How do earthworms affect the pathway of sludge bio-stabilization via vermicomposting? Science of The Total Environment, 2024, 916: 170411.
- Wang S, Yuan X, Li T, et al. Changes in soil microbe-mediated carbon, nitrogen and phosphorus cycling during spontaneous succession in abandoned PbZn mining areas. Science of The Total Environment, 2024, 920: 171018.
- Li Z, Feng C, Lei J, et al. Farmland Microhabitat Mediated by a Residual Microplastic Film: Microbial Communities and Function. Environmental Science & Technology, 2024, 58(8): 3654-3664.
- Tao K, Jensen I T, Zhang S, et al. Nitrogen and Nod factor signaling determine Lotus japonicus root exudate composition and bacterial assembly. Nature Communications, 2024, 15: 3436.
- Peng Y, Gu X, Zhang M, et al. Simultaneously enhanced autotrophic–heterotrophic denitrification in iron-based ecological floating bed by plant biomass: Metagenomics insights into microbial communities, functional genes and nitrogen metabolic pathways. Water Research, 2024, 248: 120868.


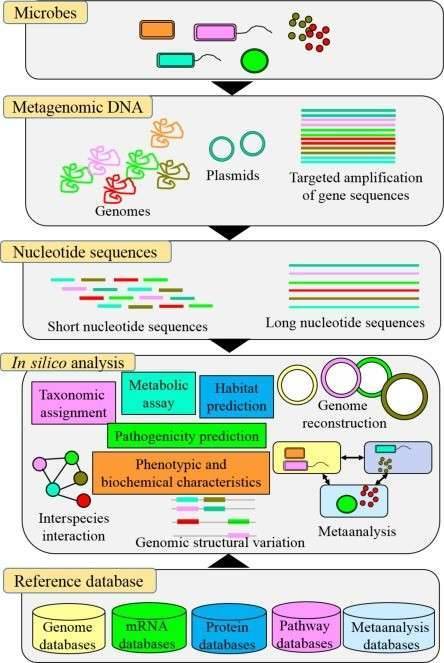
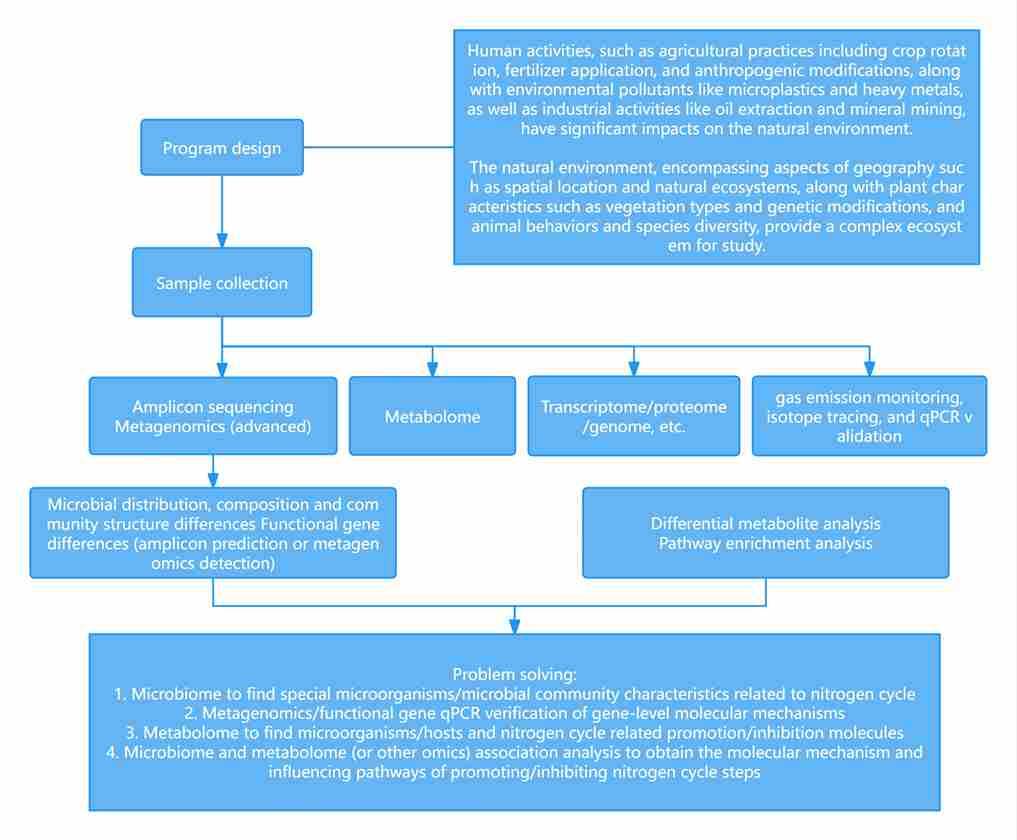 Figure 1 presents a general technical roadmap for microbial studies on the nitrogen cycle.
Figure 1 presents a general technical roadmap for microbial studies on the nitrogen cycle. Fig. 2 Potential mechanisms of nitrogen-related transformations during vermicomposting of sewage sludge
Fig. 2 Potential mechanisms of nitrogen-related transformations during vermicomposting of sewage sludge Fig.3 RDA analysis of the correlation between CNP cycling genes and soil factors
Fig.3 RDA analysis of the correlation between CNP cycling genes and soil factors Fig. 4 Gene abundance and microbial (phylum level) contributions to nitrogen and sulfur cycles in soil (n = 12) and plasticsphere (n = 12). (A) Nitrogen cycle, nitrogen fixation (black), nitrification (red), anaerobic ammonium oxidation (orange), denitrification (blue); (B) Sulfur cycle, sulfur oxidation process (blue), assimilatory sulfate reduction process (black), dissimilatory sulfate reduction and oxidation (red), sulfur fixation (orange); (C and D) Relative abundance of genes related to nitrogen cycle (C) and sulfur cycle (D) in soil and plasticsphere based on Wilcoxon rank sum test (p < 0.05); (E and F) Schematic diagram of the effect of residual microplastic film on nitrogen (E) and sulfur (F) cycle processes in farmland (*: p < 0.05)
Fig. 4 Gene abundance and microbial (phylum level) contributions to nitrogen and sulfur cycles in soil (n = 12) and plasticsphere (n = 12). (A) Nitrogen cycle, nitrogen fixation (black), nitrification (red), anaerobic ammonium oxidation (orange), denitrification (blue); (B) Sulfur cycle, sulfur oxidation process (blue), assimilatory sulfate reduction process (black), dissimilatory sulfate reduction and oxidation (red), sulfur fixation (orange); (C and D) Relative abundance of genes related to nitrogen cycle (C) and sulfur cycle (D) in soil and plasticsphere based on Wilcoxon rank sum test (p < 0.05); (E and F) Schematic diagram of the effect of residual microplastic film on nitrogen (E) and sulfur (F) cycle processes in farmland (*: p < 0.05) Figure 5. Nitrogen nutrition and signaling during root nodule symbiosis affect community assembly.
Figure 5. Nitrogen nutrition and signaling during root nodule symbiosis affect community assembly. Fig. 6. Spearman correlation analysis between functional genes and denitrification performance (a); Contribution of species to functional genes (b); Possible mechanism of ZVI and plant biomass enhancing denitrification (c)
Fig. 6. Spearman correlation analysis between functional genes and denitrification performance (a); Contribution of species to functional genes (b); Possible mechanism of ZVI and plant biomass enhancing denitrification (c)