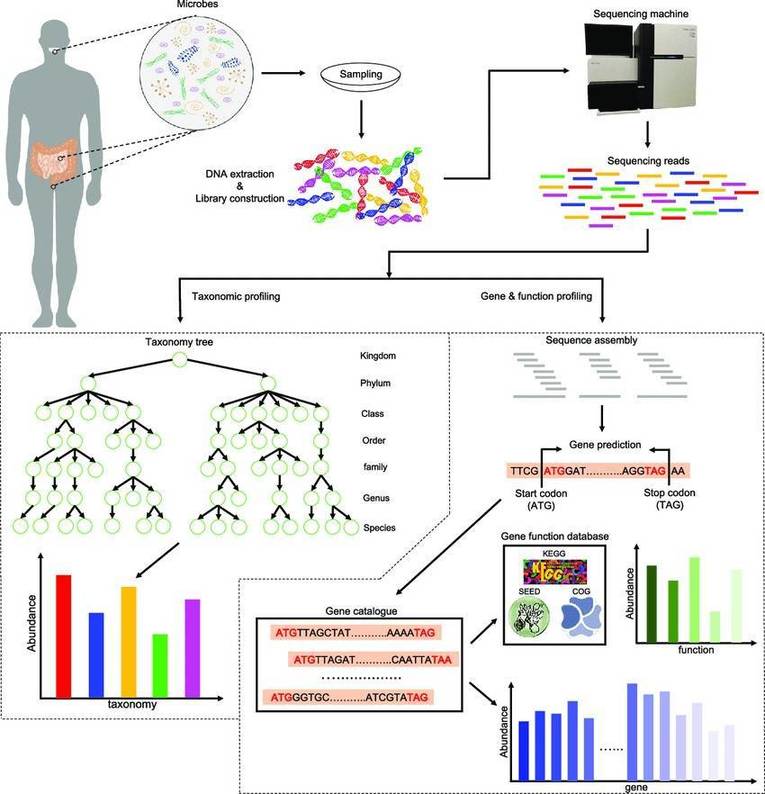
Shallow shotgun metagenome sequencing (SSMS) represents a streamlined approach to microbial community analysis by sequencing samples at a reduced depth compared to deep shotgun metagenomics. This method enhances cost-efficiency by amalgamating multiple samples within a single sequencing run and utilizing a simplified protocol that reduces reagent volumes for library preparation. SSMS effectively delivers comprehensive compositional and functional insights akin to those from deep sequencing, making it highly suitable for expansive studies that necessitate a judicious balance between the scope and resolution of microbial data.
Our Advantages:
- Cost Efficiency: SSMS is a budget-friendly alternative to deep sequencing, close to 16S rRNA costs, ideal for large-scale studies.
- High Resolution: Provides detailed microbial profiles with excellent taxonomic resolution, even with significant host DNA.
- Rapid Analysis: Delivers essential data quickly with swift turnaround times.
- Versatility: Suitable for diverse applications and sample types, from environmental to various research needs.
What is Shallow Shotgun Metagenomics
Shallow Shotgun Metagenomics is a cutting-edge sequencing approach designed to provide a broad and detailed overview of microbial communities by sequencing samples at a relatively lower depth compared to deep shotgun sequencing. Utilizing fewer sequencing reads and optimized sample preparation, SSMS offers a cost-effective alternative that maintains high resolution for microbial community analysis, delivering insights into both the taxonomic composition and functional capabilities of the microbiome.
Difference Between Shallow and Deep Shotgun Sequencing
- Shallow Shotgun Sequencing
SSMS involves sequencing samples with a lower read depth, typically ranging from 2 to 5 million reads per sample. This approach provides a cost-effective means of obtaining a comprehensive overview of microbial communities, offering reasonable taxonomic resolution, often down to the species or strain level. SSMS is particularly suitable for large-scale studies where budget constraints are significant.
In contrast, deep shotgun sequencing entails a higher read depth, generally exceeding 10 million reads per sample. This method delivers an extensive and detailed characterization of microbial communities, enabling more precise species and strain-level identification as well as comprehensive functional gene analysis. Deep sequencing is often employed in studies requiring high resolution and extensive data, such as biomarker discovery. However, it is more expensive and less feasible for large-scale projects.
- 16S rRNA Gene Amplicon Sequencing
16S rRNA gene amplicon sequencing targets specific regions of the 16S ribosomal RNA gene to classify microbial communities. While this method is cost-effective and suitable for large cohorts, it generally provides taxonomic resolution only up to the genus level and offers limited functional insights. In contrast, SSMS delivers more detailed taxonomic and functional data, comparable to deep shotgun sequencing but at a lower cost.
Applications of Shallow Shotgun Metagenome Sequencing
SSMS is a robust tool for microbial community analysis across various research areas. This method is particularly effective for:
- Microbiome Studies: Offers in-depth insights into microbial diversity and composition, especially in environments with significant host DNA contamination, where traditional deep sequencing may be prohibitively expensive.
- Epidemiological Research: Supports large-scale investigations into microbial community changes and their associations with various factors, all while keeping costs in check.
- Environmental Monitoring: Enables comprehensive analysis of microbial populations across different ecosystems, providing detailed data without the high costs associated with deeper sequencing.
- Biomedical Research: Assists in identifying microbial patterns related to different states, providing valuable information on microbial roles and interactions in various contexts.
Service Specifications
Introduction to Our Shallow Shotgun Metagenome Sequencing Services
At CD Genomics, our SSMS services provide a powerful and cost-effective solution for comprehensive microbial community analysis. Utilizing advanced sequencing technologies, we achieve approximately 3 million reads per sample, ensuring balanced depth for precise taxonomic classification and functional profiling. Our meticulous post-sequencing data processing, including contamination removal and mapping against curated databases, delivers high-resolution microbial profiles. The resulting bioinformatics analysis offers detailed taxonomic and functional insights, enhanced by interactive visualizations and advanced diversity metrics, tailored to support a wide range of research needs.
Shallow Shotgun Metagenome Sequencing Workflow

Technical Parameters
- Read Depth: Typically 2 to 5 million reads per sample.
- Platform: Illumina NextSeq or equivalent.
- Read Length: 150 bp paired-end reads.
- Quality Control: Rigorous internal quality checks throughout the workflow.
Note: The above content includes only a portion of the bioinformatics analysis. For more information or to customize the analysis, please contact us directly.
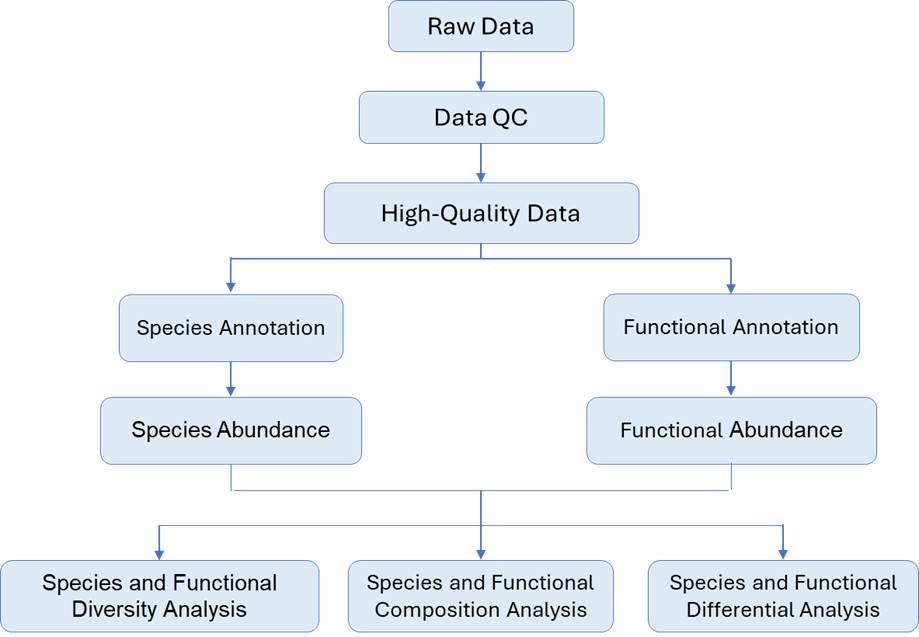
Bioinformatics Analysis
Our bioinformatics services include:
- Quality Control: We provide comprehensive metrics on sequencing quality, encompassing parameters such as read length and error rates.
- Taxonomic Profiling: Our services enable the precise identification and quantification of microbial taxa down to the species or strain level.
- Functional Analysis: We assess the functional gene content and associated pathways using established databases like KEGG and SEED
- Diversity Analysis: We offer both alpha and beta diversity metrics to elucidate community composition and variability.
- Custom Analysis: We design tailored analysis packages to meet specific research needs, including comparative and differential analyses.
Sample Requirement
- 1.8 < OD260/280 < 2.0, no degradation or contamination.
- Bacterial Draft Genome: Total DNA ≥ 500 ng, concentration ≥ 10 ng/μL
- Bacterial Complete Genome: Total DNA ≥ 20 μg, concentration ≥ 100 ng/μL
Note: If you wish to obtain more accurate and detailed information regarding sample requirements, please feel free to contact us directly.
Deliverables
- Raw sequencing data (FASTQ)
- Trimmed and stitched sequences (FASTA)
- Quality-control dashboard
- Statistic data
- Your designated bioinformatics result report
Partial results of our shallow shotgun metagenome sequencing service are shown below:
Please feel free to reach out if you have any further inquiries or require additional information.




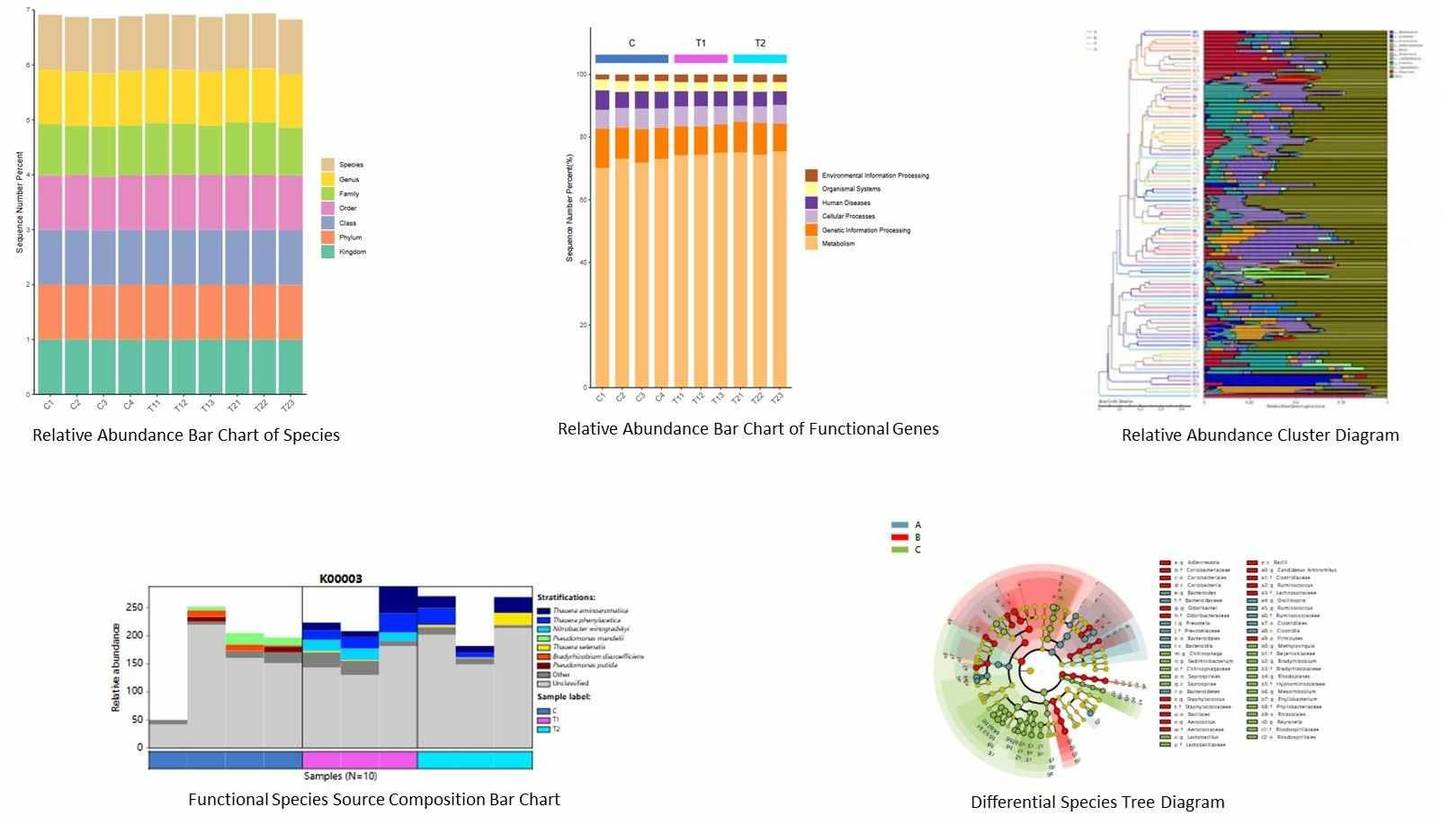

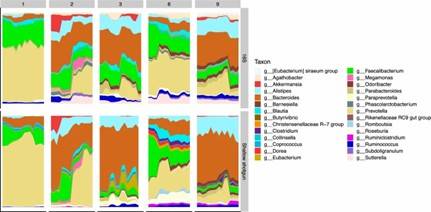 Fig 1. Area plots depicting individualized microbiome profiles at the genus level for the five subjects across their respective sampling periods on the x-axis (days).
Fig 1. Area plots depicting individualized microbiome profiles at the genus level for the five subjects across their respective sampling periods on the x-axis (days).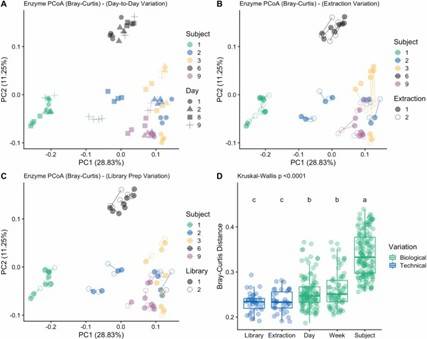 Fig 2. Principal coordinates analysis (PCoA) plot showing the Bray–Curtis dissimilarity of KEGG Enzyme profiles between all samples from SS sequencing.
Fig 2. Principal coordinates analysis (PCoA) plot showing the Bray–Curtis dissimilarity of KEGG Enzyme profiles between all samples from SS sequencing.