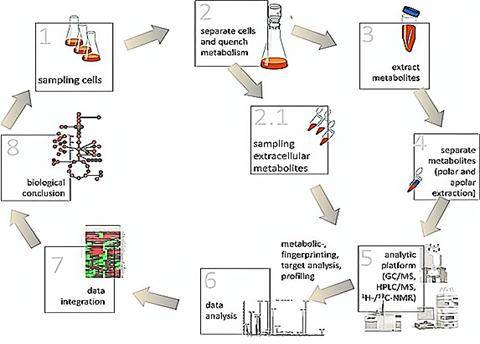
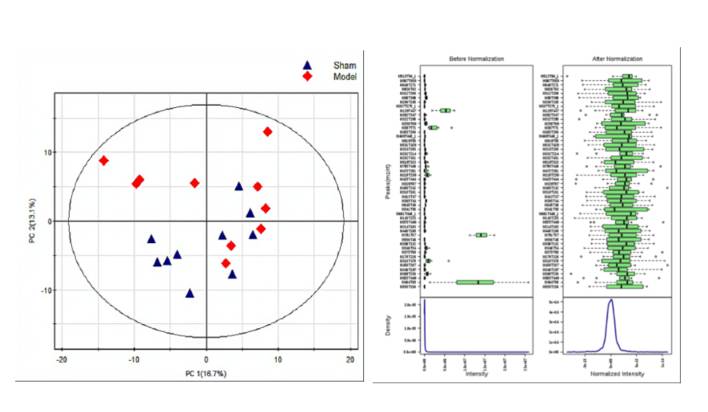
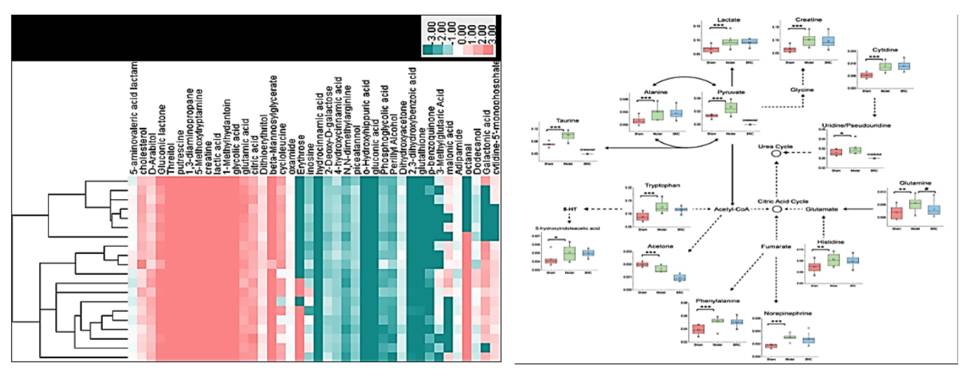
Microbial metabolomics employs tandem mass spectrometry or nuclear magnetic resonance techniques in concert with bioinformatics tools. Through these methods, qualitative and quantitative analyses of microbial metabolites are conducted offering insights into their physiological states. Low molecular weight metabolites, hormones, and signaling molecules generated throughout the life cycle or in specific physiological periods of microbes are systematically profiled and quantified using metabolomics. With the rapid development of novel analytical techniques, microbial metabolomics has garnered considerable attention for providing a more complete understanding not only of various metabolic pathways network but also of the mechanisms of interaction between microbes and their hosts. In adopting tandem mass spectrometry techniques coupled with advanced bioinformatics algorithms and tools, CD Genomics offers assistance in accurately analyzing and quantifying microbial metabolites. This provides invaluable insight into the physiological state of the microbes being examined.
Our Advantages:
- High resolution and detection sensitivity
- Analysis speed, peak capacity
- High completion rate and high accuracy
- Complete test report with user-friendly results
What Is Microbial Metabolomics?
The term metabolome, from which the field of metabolomics is derived, signifies the ensemble of all low molecular weight metabolites (this includes metabolic intermediates, hormones, signaling molecules, and secondary metabolites) within a biological cell or organism during a specific physiological phase. As the core offering an interconnection between cellular changes and phenotypes, the metabolome provides direct insight into the physiological status of a cell.
Microbial metabolomics mainly studies the analysis of intracellular or extracellular primary metabolites, signaling molecules, hormones and secondary metabolites. The ultimate goal of microbial metabolomics is to understand the physiological state of microorganisms through qualitative and quantitative analysis of microbial metabolites. In addition, the data derived from metabolomics are clearly different from those derived from genomics (what may happen), transcriptomics (what can happen), and proteomics (what happens on the basis of what happens), but rather reflect what has already happened in the cell. Microbial metabolomics can provide accurate information about the actual physiological state of microorganisms or reflect their effects on host metabolism.
Analytical Tools of Microbial Metabolomics
In the past two decades, research in microbial metabolomics has witnessed an exponential surge, with tandem mass spectrometry (MS) commanding the landscape as the most prevalent analytical methodologies. The fundamental principle of MS involves ionization of the individual components within a sample at an ion source, thereby producing ions with varying mass-to-charge ratios. By leveraging an accelerating electrical field, these ions are generated into an ion beam and directed into a mass analyzer. Within the analyzer, electric and magnetic fields induce an opposing velocity dispersion, focusing them into a mass spectrum to ascertain their mass (mass-to-charge ratio), which is then quantified or semi-quantified in response to the intensity of the mass-to-charge ratio. Mass spectrometry (MS), especially tandem MS, with its high sensitivity and separated efficiency, provides an ideal platform for both non-targeted and targeted metabolomics studies, offering additionally the advantages of convenient operation, robustness, and cost-effectiveness.
Applications of Microbial Metabolomics
Microbial metabolomics, having been widely employed in studies focusing on functional genes, microbial identification, metabolic pathways, antibiotic resistance, industrial biotechnology, synthetic biology, gut-microbial-metabolism related diseases, and enzyme discovery, is becoming more prevalent. Moreover, this advantageous field has also begun to draw the attention of scientists towards the potential for its usage in astrobiology-related biomarker detection and extraterrestrial life exploration.
Microbial Metabolomics Analysis of the Gut Microbiota
The human intestinal tract hosts an expansive range of microbes, collectively termed as the gut microbiota. These microbial communities encompass between 500 and 1000 distinctive bacterial species, with a cumulative genetic content exceeding humans by over a hundredfold. The gut microbiota notably impacts human functionalities by participating in pivotal physiological processes such as digestion, absorption, metabolism, and immune responses, thereby bearing a significant correlation with human health. Contemporary research methodologies in gut microbiota studies primarily encompass the use of 16S rRNA high-throughput sequencing for microbial diversity, along with metabolomics and proteomics. The rapid advancement in gut microbiota research analyses has been effectively propelled by the advent of systems biology. Acquire a comprehensive understanding of our specialized integrated service schemes dedicated to gut microbiome research.
The myriad of symbiotic microorganisms within the human digestive tract, along with their metabolic byproducts, have been convincingly established as playing significant roles in the pathogenesis of diverse diseases. With the advancements in multi-omics technologies, specifically the combined analytical approach of gut microbiome and metabolomics, researchers have identified the continuous cycle of "Gut Microbiota-Small Molecule Metabolites-Biological Function" as a principal core pathway through which gut microbiota exert their effects. This finding holds significant applicative value in the fields of the gut-brain axis, liver-gut axis, metabolic diseases, neurologic diseases, and many more.
The gut microorganisms generate a myriad of metabolites, such as Trimethylamine-N-oxide, short-chain fatty acids, long-chain fatty acids, 4-ethylphenylsulfate, indoles, polyamines, retinoic acids, bile acids, flavonoids, and N-acyl amides among others. The development of novel, non-targeted metabolomics technologies has made it possible to characterize the gut microbiota and its metabolites that are associated with human disease.
A precise detection and analysis of the intestinal microbiota and host metabolites, coupled with the tracking of distinct changes and variations in the gut microbial metabolic profiles, can provide a more direct perspective of the "gut microbiota-host" metabolic interplay. This holds significant implications for further understanding the physiological or pathological mechanisms underlying these complex interactions.
Service Specifications
Introduction to Microbial Metabolomics Service
In recent years, with the rapid development of new analytical technologies, CD Genomics has been dedicated to providing microbial metabolomics analysis based on tandem mass spectrometry technology, in conjunction with bioinformatics analytical tools, aiming to understand the networks of various microbial metabolic pathways. We have established metabolomics research platforms specifically targeting primary and secondary metabolites produced by microbes.
What metabolites can we analyze?
Our platforms enable the analysis of a wide array of microbial metabolic products including short-chain fatty acids, long-chain fatty acids, 4-ethylphenylsulfate, indoles, polyamines, retinoic acids, bile acids, neurotransmitters, trimethylamine N-oxide, amino acids, vitamins, and sugars etc.
Depending on your research objectives, we utilize distinct sample treatment protocols to efficiently extract microbial metabolites for analysis. This approach not only facilitates the effective addressal of your biological inquiries, but also enhances the identification and quantification accuracy of the metabolites of interest.
Sample Preparation
In order to comprehensively analyze metabolites, the sample extraction methods should satisfy several criteria: maximize metabolite extraction, maintain unbiased representation, and neither alter nor compromise the physical or chemical characteristics of the metabolites. In the practical execution of experiments, we implement a pre-experimental phase for your samples. During this phase, we employ a variety of metabolite extractions, subsequently analyzing each of the resulting samples using mass spectrometry. The raw data obtained from the mass spectrometry analysis of varied extraction methods is then subjected to in-depth bioinformatics analysis. The identified metabolites are matched with the metabolites of your research interest. Based on the results from this matching process, we then select one or two appropriate extraction methods for the analysis and processing of subsequent official batch samples.
Bioinformatics Analysis
In the realm of metabolome investigation, large volumes of data related to metabolic analysis are generated via high-throughput methods necessitating the employment of bioinformatic strategies for data handling.
The CD Genomics bioinformatics analysis team adeptly conducts comprehensive and in-depth mining of metabolome data. Initiating from raw mass spectrometry data, tasks such as peak alignment, retention time correction, and peak area extraction are diligently executed. Metabolite structural identification is pursued via precise mass number matching and secondary mass spectrometry matching, by harnessing databases. For comparative studies involving two groups, ions peaks where both groups have >50% missing values are omitted. Subsequently, data normalization is carried out via auto-scaling or UV methods. Leveraging tools such as MetaboAnalyst and SIMCA-P software, our team embarks on multidimensional and unidimensional statistical analyses that include PCA, PLS-DA, OPLS-DA, and pathway enrichment analysis. This meticulous and comprehensive approach to metabolome study ensures the provision of accurate results.
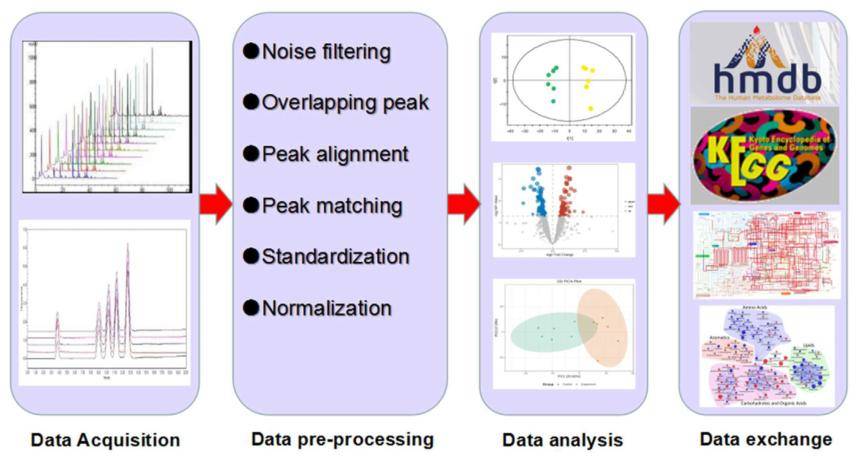 Microbial metabolomics: From novel technologies to diversified applications. (Dongyuan Ye et al., 2022)
Microbial metabolomics: From novel technologies to diversified applications. (Dongyuan Ye et al., 2022)



 Microbial metabolomics: From novel technologies to diversified applications. (Dongyuan Ye et al., 2022)
Microbial metabolomics: From novel technologies to diversified applications. (Dongyuan Ye et al., 2022)