We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy

Extrachromosomal circular DNA (eccDNA) is a significant genomic element, frequently harboring oncogenes and regulatory components like promoters and enhancers. This makes eccDNA an essential focus in cancer research, where it is linked to key processes such as oncogene amplification, regulation of gene expression, genomic rearrangements, and tumor heterogeneity. As research progresses, our understanding of eccDNA's biological functions and its role in cancer has become more nuanced.
The identification and analysis of eccDNA have advanced with the development of various analytical algorithms and experimental techniques. Some of the most widely used methods include the AmpliconArchitect (AA) algorithm, CReSIL, Circle_finder, Circle-seq, and 3SEP. Despite these advances, eccDNA's structural complexity and size variability pose significant challenges in selecting the optimal detection method for different types of studies.
Current evaluations of eccDNA detection strategies tend to be narrow in focus, often assessing only one aspect, such as accuracy or computational efficiency, while simplifying the simulations used to test these methods. This limited scope often fails to account for the complexities of real-world sequencing data, highlighting the need for more comprehensive and realistic benchmarks to guide method selection in eccDNA research.
Take the Next Step: Explore Related Services
Learn More
To address the challenges mentioned above, a research team led by Qu Kun and Guo Chuang from the University of Science and Technology of China published an article titled "Comparative analysis of methodologies for detecting extrachromosomal circular DNA" in Nature Communications.
The team analyzed seven algorithms for identifying eccDNA in sequencing data using seven simulated datasets, evaluating their performance in terms of accuracy, identity recognition, repeatability, and computational resource consumption. Additionally, they compared the detection efficiency of seven experimental library preparation methods for different eccDNA types using 21 real sequencing datasets. This comparative study highlighted the most effective methods for analyzing short-read (SR) and long-read (LR) sequencing data enriched with eccDNA, emphasizing the differences in detection efficiency across various experimental methods. It provides valuable guidance for researchers in selecting appropriate methodologies, thus advancing the development of efficient new methods for eccDNA detection.
To assess the performance of different analysis pipelines in eccDNA detection, the research team developed a Python script to simulate eccDNA datasets, incorporating factors such as length distributions, chromosomal origins, and chimeric eccDNA. This simulation allowed the evaluation of various algorithms for SR and LR sequencing data, providing insights into their strengths and limitations. The study also compared the effectiveness of several experimental methods, including Circle-Seq, 3SEP, whole-genome sequencing (WGS), and ATAC-Seq, in detecting eccDNA across different sample types and sequencing depths.
Through these evaluations, the research team identified key factors influencing eccDNA detection efficiency, such as enrichment methods and sequencing depth, which are crucial for optimizing detection strategies in cancer research and diagnostics. The following sections will delve into the detailed findings from the benchmarking of eccDNA analysis pipelines and the impact of various experimental approaches on eccDNA detection.
Research Design
To evaluate the performance of different analysis pipelines in eccDNA detection, the research team developed a Python script to generate simulated eccDNA datasets. This script inferred length distributions, chromosomal origins, and the proportion of chimeric eccDNA from existing data, creating mixed datasets containing both circular DNA (true positives) and linear DNA (true negatives). Additionally, the script simulated the rolling circle amplification (RCA) process. In total, seven simulated datasets were generated, each containing 10,000 circular DNA sequences and 10,000 linear DNA sequences at 50X coverage.
The research team assessed 11 configurations of seven algorithms, including Circle-Map, Circle_finder (using bwa-mem-samblaster and microDNA.InOne.sh), ECCs_plorer, and ecc_finder (using map-sr and asm-sr) for SR sequencing data analysis. For LR sequencing data analysis, they evaluated CReSIL, eccDNA_RCA_nanopore, NanoCircle, and ecc_finder (using map-ont and asm-ont). Performance metrics included F1 scores and base pair differences between identified eccDNA and the simulated eccDNA.
For the experimental method evaluation, the research team selected Circle-Seq (SR and LR), 3SEP (SR and LR), whole-genome sequencing (WGS, SR and LR), and ATAC-Seq (SR) to assess eccDNA detection efficiency under various length and copy number conditions.
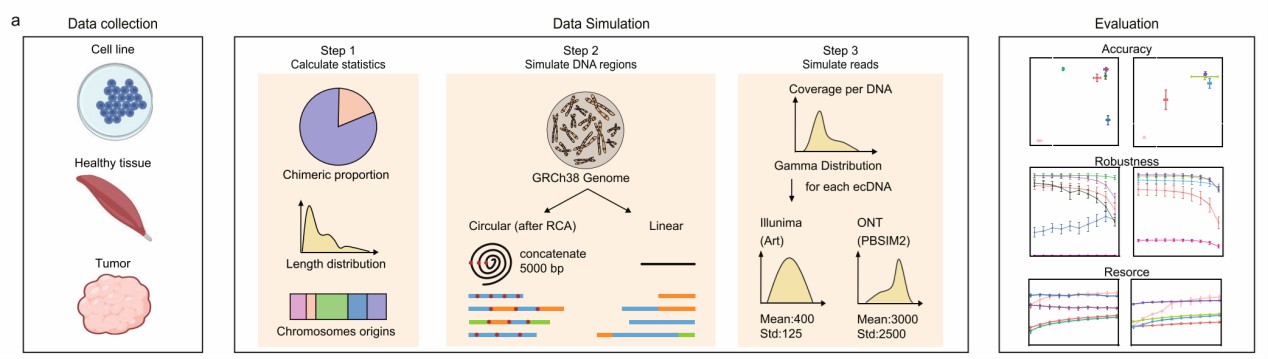 Schematic overview of the benchmarking workflow (Gao et al., 2024)
Schematic overview of the benchmarking workflow (Gao et al., 2024)
Evaluation of Different Analysis Algorithms for eccDNA Identification
At a simulated sequencing depth of 50×, the research team evaluated the performance of each analysis algorithm for eccDNA identification. The results revealed that Circle_finder (bwa-mem-samblaster) and Circle-Map outperformed other methods for SR sequencing data, with F1 scores of 0.912 and 0.908, respectively. For LR sequencing data, CReSIL performed the best with an F1 score of 0.918 and a base pair difference of 4.160 bp.
Subsequently, the research team assessed the performance of each algorithm at different sequencing depths. For SR sequencing data, Circle_finder (bwa-mem-samblaster) and Circle-Map consistently achieved the highest F1 scores at all sequencing depths. When the sequencing depth decreased from 50× to 5×, Circle-Map and Circle_finder (microDNA.InOne.sh) maintained stable base pair differences. ecc_finder showed the lowest F1 scores across all sequencing depths. For LR sequencing data, CReSIL exhibited the highest F1 scores when the depth exceeded 10×, while eccDNA_RCA_nanopore demonstrated superior performance at depths below 10×.
In addition to sequencing depth, the research team also investigated the impact of chimeric DNA on eccDNA identification performance. For SR sequencing data analysis, changes in the proportion of chimeric DNA did not affect the eccDNA identification recall rate for Circle-finder (bwa-mem-samblaster), Circle-Map, and ecc_finder (map-sr), but it did affect ECCs_plorer. For LR sequencing data analysis, most algorithms maintained consistent recall rates when identifying both simple and chimeric eccDNA.
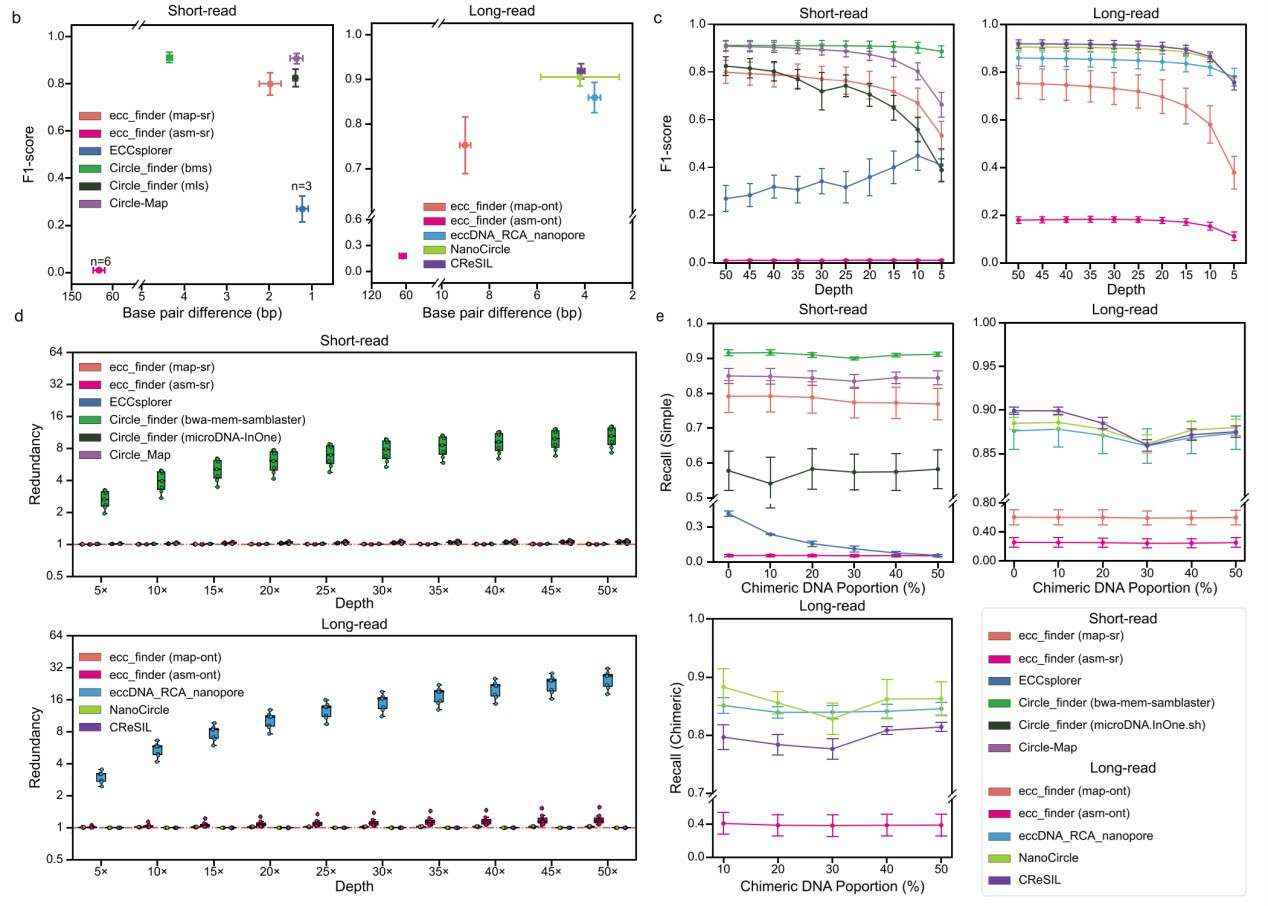 Assessment of analysis pipelines in eccDNA identification (Gao et al., 2024)
Assessment of analysis pipelines in eccDNA identification (Gao et al., 2024)
Based on the analysis, Circle_finder (bwa-mem-samblaster) and Circle-Map are the most suitable algorithms for analyzing eccDNA-enriched SR sequencing data, though Circle_finder tends to generate redundant results. CReSIL outperforms other algorithms for analyzing eccDNA-enriched LR sequencing data, providing higher detection accuracy and smaller base pair differences.
Take the Next Step: Explore Related Services
Impact of Enrichment Steps on eccDNA Identification
The research team next assessed the eccDNA detection efficiency based on the number of eccDNA detected per Gb of data. The results revealed that methods employing the RCA step significantly outperformed those without RCA in terms of eccDNA detection efficiency. Furthermore, a positive correlation was observed between genomic copy number and the coverage of overlapping eccDNA.
Further analysis of eccDNA length distribution and chromosomal origins showed that over 97% of eccDNA detected by enrichment methods were shorter than 10 kb, whereas a higher proportion of eccDNA detected by non-enrichment methods exceeded 10 kb. Except for 3SEP-SR and WGS-SR, the eccDNA density (eccDNA detected per million base pairs) for most methods showed a significant positive correlation with the density of protein-coding genes on chromosomes.
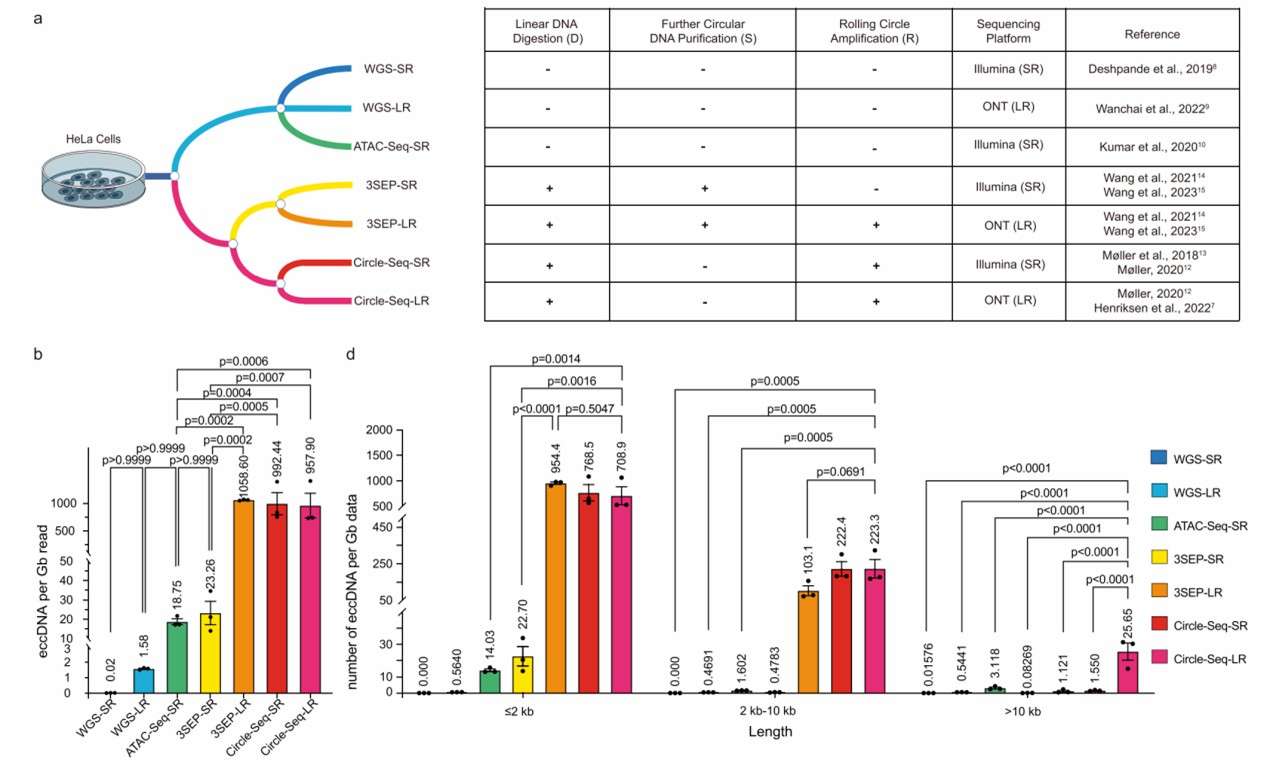 Impact of eccDNA enrichment operations on eccDNA identification (Gao et al., 2024)
Impact of eccDNA enrichment operations on eccDNA identification (Gao et al., 2024)
Detection Efficiency of ecDNA by Different Experimental Methods
EcDNA that overlaps with amplification regions is classified as ecDNA, while eccDNA outside of these regions is classified as non-ecDNA. The study found that Circle-Seq-SR, Circle-Seq-LR, and 3SEP-LR identified a higher average number of ecDNA per Gb of data, while WGS-SR, WGS-LR, and ATAC-Seq-SR identified a significantly higher proportion of ecDNA within the detected eccDNA.
The research team further analyzed the detection efficiency of ecDNA and non-ecDNA across different length categories (≤2kb, 2-10kb, >10kb). The results showed that 3SEP-LR exhibited the highest efficiency for detecting ecDNA and non-ecDNA of lengths ≤2kb. Circle-Seq-SR performed the best for detecting ecDNA in the 2-10kb range, while Circle-Seq-LR outperformed other methods in detecting ecDNA greater than 10kb.
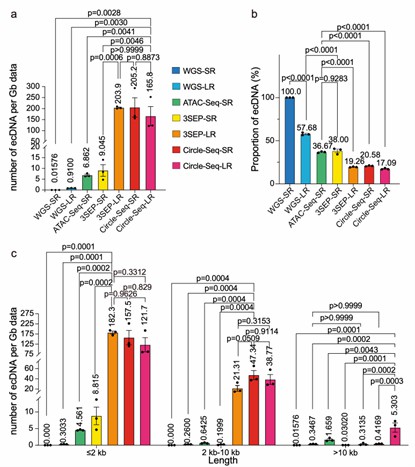 Detection efficiency of ecDNA by 7 experimental methods (Gao et al., 2024)
Detection efficiency of ecDNA by 7 experimental methods (Gao et al., 2024)
Additionally, the ecDNA profiles detected by different experimental methods showed significant heterogeneity. These methods identified eccDNA with notable differences in length, oncogene composition, and the inclusion of gene repetitive elements. Therefore, when comparing the results across different studies, it is essential to consider the experimental methods used.
This study offers a comprehensive evaluation of seven algorithms and experimental methods for eccDNA detection, providing valuable insights into their performance across different sequencing strategies. The analysis reveals important differences in the effectiveness of these methods for various sequencing types. For SR sequencing, the algorithms Circle_finder (bwa-mem-samblaster) and Circle-Map emerged as the most reliable for detecting eccDNA with high precision, demonstrating their robustness and efficiency in accurately identifying circular DNA fragments in high-throughput datasets. In contrast, for LR sequencing, CReSIL stood out as the superior method, delivering enhanced accuracy and minimizing error rates, making it the go-to choice for more complex genomic landscapes where longer reads are required to capture the full diversity of eccDNA.
When examining experimental methods, Circle-Seq-LR was found to be particularly adept at detecting larger eccDNA fragments, showing greater sensitivity and efficiency for longer circular DNA sequences, which are often challenging to identify with traditional methods. On the other hand, 3SEP-LR demonstrated exceptional performance in detecting smaller eccDNA species, showcasing its capability to efficiently identify short eccDNA fragments that are typically underrepresented in other detection strategies.
These findings underscore the critical importance of selecting the appropriate combination of algorithms and experimental techniques depending on factors such as the length of eccDNA fragments and the sequencing depth. This nuanced approach ensures more accurate and comprehensive eccDNA detection, contributing to a deeper understanding of their role in genomic instability, cancer progression, and tumor heterogeneity.
Ultimately, this study serves as a crucial reference for researchers in the field of eccDNA and cancer genomics. By guiding the selection of the most effective tools and methodologies for eccDNA detection, the research paves the way for further exploration into the functional implications of eccDNA in various diseases, particularly in the context of oncogene amplification and genetic variability in tumors. This enhanced detection framework could lead to more targeted therapeutic strategies, offering potential breakthroughs in cancer diagnosis and treatment.
Reference

CD Genomics is transforming biomedical potential into precision insights through seamless sequencing and advanced bioinformatics.
We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy