eccDNA Validation Service By PCR And Sanger Sequencing
Why is eccDNA Validation Necessary
eccDNA validation is crucial to guaranteeing the precision and dependability of research focusing on extrachromosomal circular DNA. It enhances knowledge of the biological importance and potential functional ramifications of eccDNA by providing important information regarding the structural and sequence integrity of eccDNA.
Circular structure verification: eccDNA validation confirms that the identified DNA molecules do, in fact, have a circular structure. That the DNA molecules are not linear pieces or sequencing or sample preparation artifacts is confirmed.
Confirmation of Junction Sites: It is essential to confirm the junction sites of eccDNA in order to make sure that the circular DNA molecules are created using the proper processes, like DNA replication or recombination. It contributes to proving the veracity of the circular DNA structures.
Elimination of Contamination: eccDNA validation aids in the detection and exclusion of any potential linear DNA or other sources of contamination that might obstruct a precise examination of the circular DNA.
High-Throughput Sequencing Reliability: Validating eccDNA using Sanger sequencing adds more assurance to the precision and dependability of high-throughput sequencing techniques utilized for eccDNA detection. It aids in the validation of the outcomes produced by next-generation sequencing methods.
eccDNA Validation Service
To confirm the circular structure of particular circular DNA uncovered during sequencing efforts, CD Genomics provides Sanger sequencing services. To confirm the sequence at the junction point, PCR amplification is carried out using reverse primer designs. To precisely determine the junction site of circular DNA, Sanger sequencing is applied to the PCR products covering the junction site. The chosen circular junction locations are more thoroughly verified by Sanger low-throughput sequencing. Additionally, it confirms that circular DNA can be correctly identified using high-throughput sequencing and analysis. It also validates the presence of several circular DNA molecules with low expression that were discovered using high-throughput sequencing.
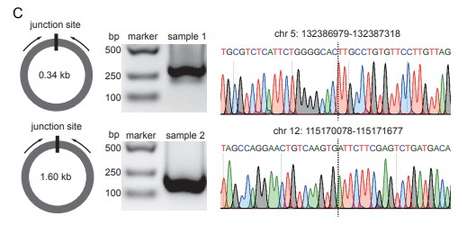 PCR & Sanger-sequencing validation of cell-free eccDNA junction site
PCR & Sanger-sequencing validation of cell-free eccDNA junction site
Workflow
1.PCR Amplification: Create unique PCR primers that specifically target the desired eccDNA sequence. To amp up the circular DNA molecule, the primers must be placed on either side of the circular junction. Perform PCR using a DNA sample that is thought to contain eccDNA and the designed primers. It is important to tune the PCR conditions for effective circular DNA amplification.
2. Gel Electrophoresis: Run the PCR results through an agarose gel to analyze them. Based on the primer locations, it is possible to estimate the predicted size of the amplified eccDNA fragment. By using a gel extraction kit and per to the manufacturer's recommendations, you can remove the amplified eccDNA band from the gel. To separate the eccDNA fragment for subsequent Sanger sequencing, this procedure is required.
3. Sanger Sequencing and analysis:Perform Sanger sequencing of the gel-extracted eccDNA fragment using appropriate primers. This sequencing step confirms the sequence of the amplified eccDNA and provides information about any potential mutations or modifications. Analyze the Sanger sequencing results to validate the presence and characteristics of eccDNA, including its sequence, potential mutations, and any modifications present. By combining PCR amplification and Sanger sequencing, researchers can validate the presence and sequence of eccDNA molecules and obtain additional information about their structure and potential genetic variations.
Sample Requirements
| Sample Type |
Sample Volume |
| Body Fluid Samples |
|
| Serum |
5 mL |
| Plasma |
5 mL |
| Urine |
5-10 mL |
| Cerebrospinal Fluid (CSF) |
5 mL |
| Tissue/Cell Samples |
|
| Cells |
2 x 10^7 |
| Tissue |
200 mg |
| Genomic DNA |
>=10 µg
(Dissolved in sterile water or TE buffer, pH=8)
(OD 260/280 ratio should be between 1.7 and 2.0)
(RNA should be removed completely)
(No contamination of DNA from other individuals or species) |
Reference
-
Jingwen Fang et al,.ScanTecc classifies primary cancers via cell-free extrachromosomal circular DNA in peripheral blood medRxiv 2022
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.
