eccDNA Methylation Sequencing Service
What is eccDNA Methylation
EcDNA (extrachromosomal DNA) is used to describe DNA outside of the chromosomes that exceeds several hundred kilobases (kb) in size, while eccDNA refers to all extrachromosomal circular DNA smaller than ecDNA. Most eccDNA molecules are smaller than 25kb and are primarily distributed between 0.1-5kb. In 2019, articles published in Nature and Cell explored the structure of eccDNA and found that it carries not only complete genes but also upstream promoter and enhancer elements. EccDNA can carry intact genes, especially oncogenic driver genes frequently found in tumors. Uncontrolled expression of these genes carried by eccDNA leads to malignant tumor growth. Extrachromosomal circular DNA (eccDNA) methylation refers to the methylation of DNA molecules that exist as circular entities outside of the chromosomal DNA. The methylation status of eccDNA in the circulation is diferent and unique under specifc physiological conditions, which makes eccDNA a trace biomarker.Perform rapid sequencing and methylation analysis of eccDNA allows assessment of cancer heterogeneity, oncogene overexpression, and environmental adaptation potential.
eccDNA Methylation Sequencing
CD Genomics' circular DNA methylation sequencing technology is based on the transposase and m5C site enzymatic conversion methods to obtain methylated circular DNA. It combines high-throughput sequencing technology, specifically DNA paired-end sequencing. This technology allows for simultaneous detection of circular DNA and the methylation status of circular DNA, providing a new technical approach for in-depth research on circular DNA and the development of novel molecular biomarkers.
eccDNA Methylation Sequencing Workflow
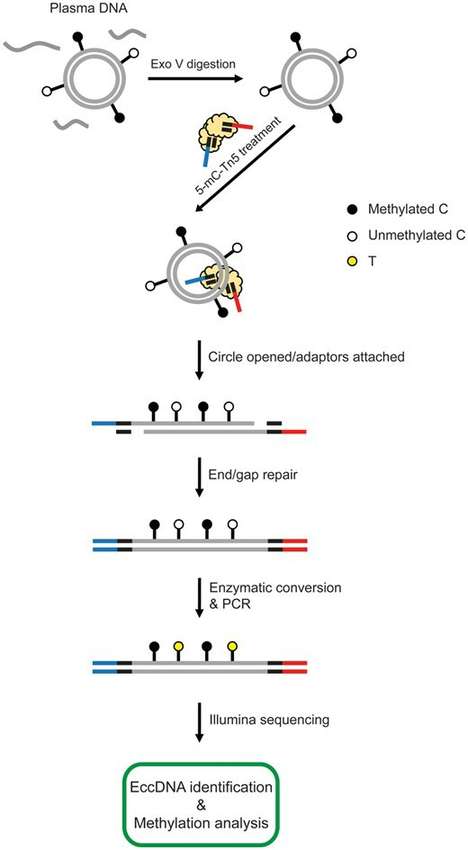
eccDNA Enrichment: The linear DNA in the sample is removed through methods such as digestion with nucleases like Exonuclease V, aiming to enrich eccDNA.
Adapter Ligation: Transposase is used to open the circular structure of eccDNA and attach adapters to both ends of the DNA fragments.
End Repair: Klenow enzyme is employed to repair the gaps at the ends of the transposed products.
C-U Conversion: A gentle enzymatic conversion method efficiently converts unmethylated cytosine (C) to uracil (U).
PCR Amplification: The converted products are subjected to PCR amplification and purification.
Sequencing: After quality inspection and purification of the library, it is ready for sequencing.
eccDNA Methylation Sequencing Data Analysis
Identification and methylation level of eccDNA.
Gene annotation of eccDNA.
Length distribution plot of eccDNA.
Statistical analysis of the methylation levels of eccDNA.
Sample Requirements
| Sample Type |
Sample Volume |
| Serum |
5 mL |
| Plasma |
5 mL |
| Urine |
5-10 mL |
| Cerebrospinal Fluid (CSF) |
5 mL |
| Tissue |
100 mg |
| cfDNA |
>=50 ng |
Application Case
Characteristics of Fetal Extrachromosomal Circular DNA in Maternal Plasma: Methylation Status and Clearance
Journal: Clinical Chemistry
Impact Factor: 8.322
Professor Yuk Ming and his team developed a sequencing approach for analyzing the methylation of circular DNA. By evaluating the levels of fetal extrachromosomal circular DNA at different postpartum time points, they investigated the in vitro clearance efficiency of fetal circular DNA. The authors also found that the methylation levels of fetal circular DNA in maternal plasma were relatively low compared to maternal circular DNA. Additionally, similar to linear fetal DNA, fetal circular DNA rapidly disappears from maternal blood after delivery. In summary, the team developed a circular DNA methylation sequencing technology based on transposase and m5C site enzymatic conversion methods. They revealed the methylation status of fetal circular DNA in maternal plasma, providing a new technical approach for in-depth research on circular DNA and the development of novel molecular biomarkers.
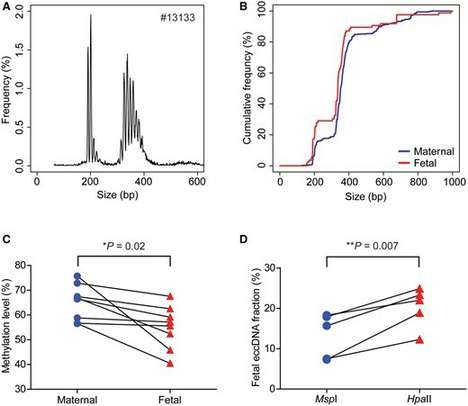 Figure 1: Length and methylation analysis of circular DNA from maternal and fetal sources.
Figure 1: Length and methylation analysis of circular DNA from maternal and fetal sources.
Reference
-
Sarah T K Sin and others, Characteristics of Fetal Extrachromosomal Circular DNA in Maternal Plasma: Methylation Status and Clearance, Clinical Chemistry, Volume 67, Issue 5, May 2021, Pages 788–796
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.
