eccDNA (circular DNA) Sequencing Service
What Is eccDNA
eccDNA (extrachromosomal circular DNA), also known as extrachromosomal circular DNA, is widely present in eukaryotic cells, including humans. They are typically derived or shed from the normal genome and exist as circular DNA molecules outside the chromosomal genome. Whole-genome sequence analysis of eccDNA in yeast, nematodes, ciliates, and mammals has revealed a range of sizes, from a few tens of base pairs to hundreds of thousands of base pairs. eccDNA not only carries intact or partial genes but also carries intergenic sequences, and some contain replication origins, enabling independent replication.
Since the discovery of eccDNA molecules in mammals and higher plants in 1965, it has been found that eccDNA is abundant in tumor cells and often carries oncogenes. It can promote tumor development by increasing the expression of oncogenes. Two articles published in Nature and Cell in 2019 regarding eccDNA in cancer further suggest that eccDNA exhibits significant chromosome open state and long-range interactions. The enhancer function in the non-coding regions of eccDNA also plays a role in the regulation of oncogene expression carried by eccDNA, further linking the epigenetic features of eccDNA to cancer. Additionally, eccDNA is associated with drug resistance, aging, tumor heterogeneity, intercellular communication, and other functions.
Mechanisms of Circular DNA Formation
Current research suggests that circular DNA is formed through various mechanisms, including chromosomal breaks, looping, or additional replication. There are two proposed mechanisms for the processing of circular DNA formation:
Intrachromatid Ectopic Homologous Recombination (HR): This mechanism involves the chromosomal ectopic homologous recombination that leads to looping and circularization of the DNA sequence.
Nonhomologous End-Joining (NHEJ): This mechanism involves the nonhomologous end-joining of chromosomal ends, resulting in circularization of the DNA.
The formation of circular DNA is a complex process, and there are likely more mechanisms yet to be discovered and explored.

Advantages of eccDNA (circular DNA) Sequencing Services
High Detection Rate of Circular DNA: Multiple methods are employed to efficiently enrich circular DNA, resulting in a high detection rate of circular DNA.
Professional Bioinformatics Analysis: A dedicated team of bioinformatics experts has developed efficient algorithms for identifying and analyzing circular DNA, meeting various in-depth data analysis needs of customers.
One-Stop Service: Customers only need to provide cells, tissues, or genomic DNA, and CD Genomics will handle the entire service process, including sample preparation, circular DNA enrichment, library preparation, sequencing, and data analysis.
Sample Requirements
| Sample Type |
Sample Volume |
| Body Fluid Samples |
|
| Serum |
5 mL |
| Plasma |
5 mL |
| Urine |
5-10 mL |
| Cerebrospinal Fluid (CSF) |
5 mL |
| Tissue/Cell Samples |
|
| Cells |
2 x 10^7 |
| Tissue |
200 mg |
| Genomic DNA |
>=10 µg
(Dissolved in sterile water or TE buffer, pH=8)
(OD 260/280 ratio should be between 1.7 and 2.0)
(RNA should be removed completely)
(No contamination of DNA from other individuals or species) |
Workflow of eccDNA Sequencing

Application Case
Case 1
Circular DNA elements of chromosomal origin are common in healthy human somatic tissue(Nature Communication,2018,IF=11.878)
This study collected skeletal muscle samples and blood samples from 8 regularly exercising individuals and 8 sedentary individuals. Cell lysis and column purification methods were used to obtain eccDNA, followed by rolling circle amplification and library preparation for sequencing. Approximately 100,000 different eccDNA sequences were identified through the analysis of sequencing data, with the majority of them being less than 25 kb in length and mainly distributed between 0.1 and 5 kb. Among the eccDNA detected in the muscle, 43,960 were highly reliable. In the white blood cells, 6,253 eccDNA sequences were highly reliable, and there were no differences observed between the exercise group and the sedentary group. Furthermore, the distribution of eccDNA on the genome was explored to determine specific chromosomal origins and locations of eccDNA. Combined analysis with RNAseq data revealed gene preferences in eccDNA formation.
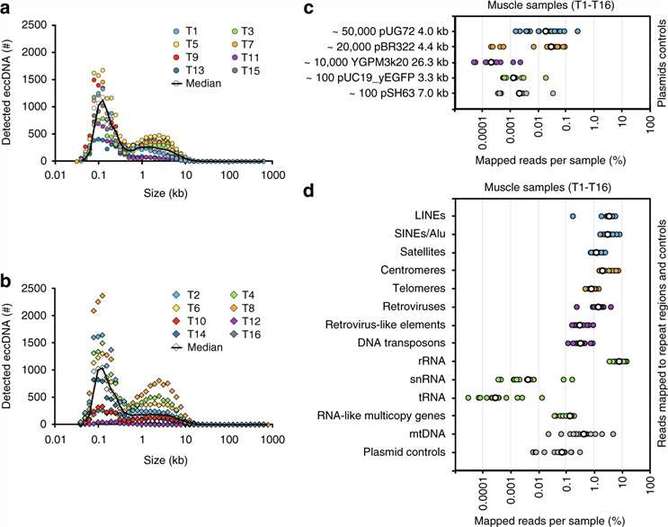 Figure 1 the reads distribution of eccDNA.
Figure 1 the reads distribution of eccDNA.
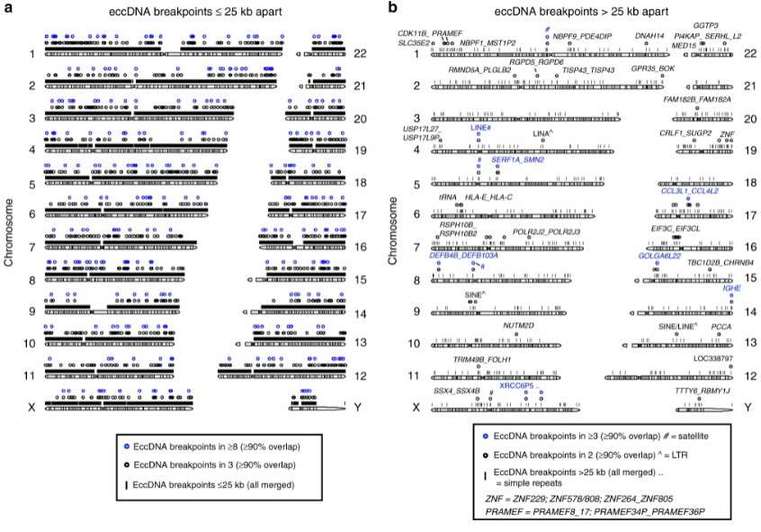 Figure 2 Genomic distribution of eccDNA
Figure 2 Genomic distribution of eccDNA
Case 2
Circular ecDNA promotes accessible chromatin and high oncogene expression(Nature,2019,IF=43.07)
The study employed integrated imaging, long-range optical mapping, epigenetics techniques, and high-throughput sequencing to provide the first comprehensive analysis of the structure and basic functions of ecDNA harboring oncogenes. By integrating RNA-seq and WGS data from tumor cell lines and TCGA clinical tumor samples, it was demonstrated that highly expressed cancer genes are encoded by ecDNA. ChIP-seq and immunofluorescence techniques were used to analyze the occurrence of active histone modifications (H3K4me1, H3K27ac) and repressive histone modifications (H3K9me3, H3K27me3) in active cancer cells, revealing a high abundance of active histone modifications and a small amount of repressive histone modifications on ecDNA. ATAC-seq and MNase-seq were employed to demonstrate that ecDNA chromatin is highly accessible and more open than DNA on nuclear chromosomes. By using the PLAC-seq (Hi-ChIP) technique, which analyzes the spatial structure of chromatin, it was discovered that ecDNA can form 3D functional domains, suggesting that ecDNA can exert long-range spatial regulation.
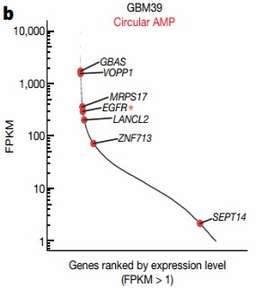 Figure 1 Highly expressed cancer genes on eccDNA
Figure 1 Highly expressed cancer genes on eccDNA
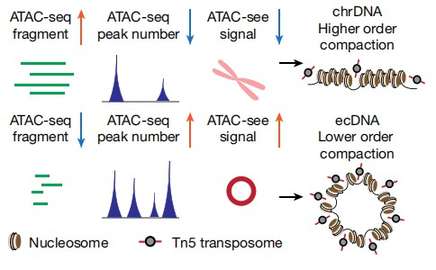 Figure 2 Depiction and interpretation of integrated technologies to assess chromatin compaction.
Figure 2 Depiction and interpretation of integrated technologies to assess chromatin compaction.
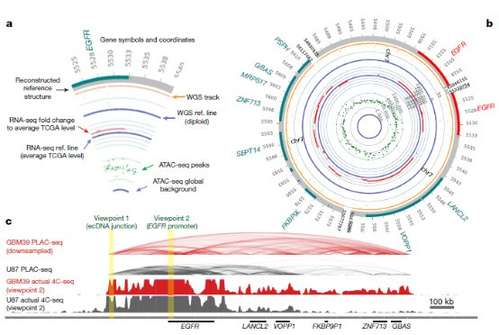 Figure 3 ircularization of ecDNA enables distal DNA interaction
Figure 3 ircularization of ecDNA enables distal DNA interaction
References
- Sin STK, Jiang P, Deng J, Ji L, Cheng SH, Dutta A, Leung TY, Chan KCA, Chiu RWK, Lo YMD. Identification and characterization of extrachromosomal circular DNA in maternal plasma. Proc Natl Acad Sci U S A. 2020 Jan 21;117(3):1658-1665
- Yang H, He J, Huang S, Yang H, Yi Q, Tao Y, Chen M, Zhang X, Qi H. Identification and Characterization of Extrachromosomal Circular DNA in Human Placentas With Fetal Growth Restriction. Front Immunol. 2021 Dec 21;12:780779
- Wei Xiang, Tongyi Zhang, Song Li, Yunquan Gong, Xiaoqing Luo, Jing Yuan, Yaran Wu, Xiaojing Yan, Yan Xiong, Jiqin Lian, Guangyu Zhao, Changyue Gao, Liang Kuang, Zhenhong Ni. cir-DNA sequencing revealed the landscape of extrachromosomal circular DNA in articular cartilage and the potential roles in osteoarthritis. bioRxiv 2022.01.27.477985
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.

