We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy

Extrachromosomal circular DNA (eccDNA) refers to a class of circular DNA molecules that exist independently of chromosomes. It is commonly found in various eukaryotic organisms, including humans, and plays a role in diverse biological processes, such as regulating gene expression, cell proliferation, DNA repair, and tumorigenesis. These molecules are less susceptible to nuclease degradation, contributing to their structural stability. Currently, the primary techniques used to study eccDNA include DNA fluorescence in situ hybridization (FISH), bulk whole-genome sequencing (WGS), and Circle-Seq. The first two methods primarily detect abundant eccDNA present in large genomic regions, while Circle-Seq offers broader detection capabilities but requires a larger sample size for sequencing.
As a result, there is an increasing need for innovative and scalable approaches to study eccDNA in individual cells or nuclei isolated from tissue biopsies. This would enable a deeper understanding of the biological roles of eccDNA and uncover its heterogeneity in tumor samples derived from patients.
A team of researchers from the Karolinska Institute in Sweden recently published a study titled "scCircle-seq unveils the diversity and complexity of extrachromosomal circular DNAs in single cells" in Nature Communications. The team developed and validated the single-cell application of Circle-Seq, named scCircle-seq, which can be applied to both fixed and unfixed cells or nuclei, including those extracted from tumor biopsy tissues.
Their study revealed that most eccDNAs are highly variable between cells of the same type and tend to form in amplified genomic regions. Applying scCircle-seq to tumor samples from three different cancer types, they discovered tumor-specific eccDNA landscapes and subclonal populations with distinct eccDNA genomic patterns.
In summary, scCircle-Seq is an easily scalable, straightforward and versatile method that promises to reveal the biological complexity and heterogeneity of eccDNA in cancer.
Take the Next Step: Explore Related Services
Learn More
In this study, the team demonstrated the power of scCircle-seq in characterizing eccDNA diversity and distribution at the genomic and cellular levels. They applied the method to a range of cell lines and patient-derived tumor samples, revealing significant variability in eccDNA profiles both within and between cell types. Moreover, the study explored the relationship between eccDNA and gene expression, showing how certain oncogenes on eccDNA may influence cellular transcription. By leveraging scCircle-seq's ability to profile eccDNA in single cells, the researchers were able to uncover unique eccDNA signatures that could be used for cell type classification and may offer insights into the epigenomic characteristics of different cell types and cancer subtypes. The findings highlight the potential of scCircle-seq to advance our understanding of eccDNA's role in normal and disease contexts, including its implications for cancer diagnostics and therapeutic strategies.
Development and Validation of scCircle-seq
To develop scCircle-seq, the researchers introduced an additional DNA nick repair step to the Circle-Seq protocol, enhancing eccDNA detection efficiency. They also designed a universal workflow applicable to both live and fixed cells or nuclei sorted in multi-well plates or single tubes.
For scCircle-seq data analysis, the team adapted a bioinformatics pipeline previously used for large-scale Circle-Seq data. This pipeline initially identifies genomic regions with high sequencing coverage, referred to as circular DNA-producing regions (CPRs). It then pinpoints chimeric junctions within each CPR by detecting overrepresented discordant and split reads mapped to these regions. The specificity of scCircle-seq was tested and found to align with expectations.
The team applied scCircle-seq to five distinct cell lines, including four cancer-derived lines (HeLa, K562, Colo320DM, and PC3) and one immortalized normal cell line (293T). The number of identified CPRs and their corresponding genomic coverage varied among single cells and cell lines. These findings are consistent with previously published Circle-Seq data, supporting the conclusion that most eccDNA exhibits significant cell-to-cell variation and is inherited randomly during cell division.
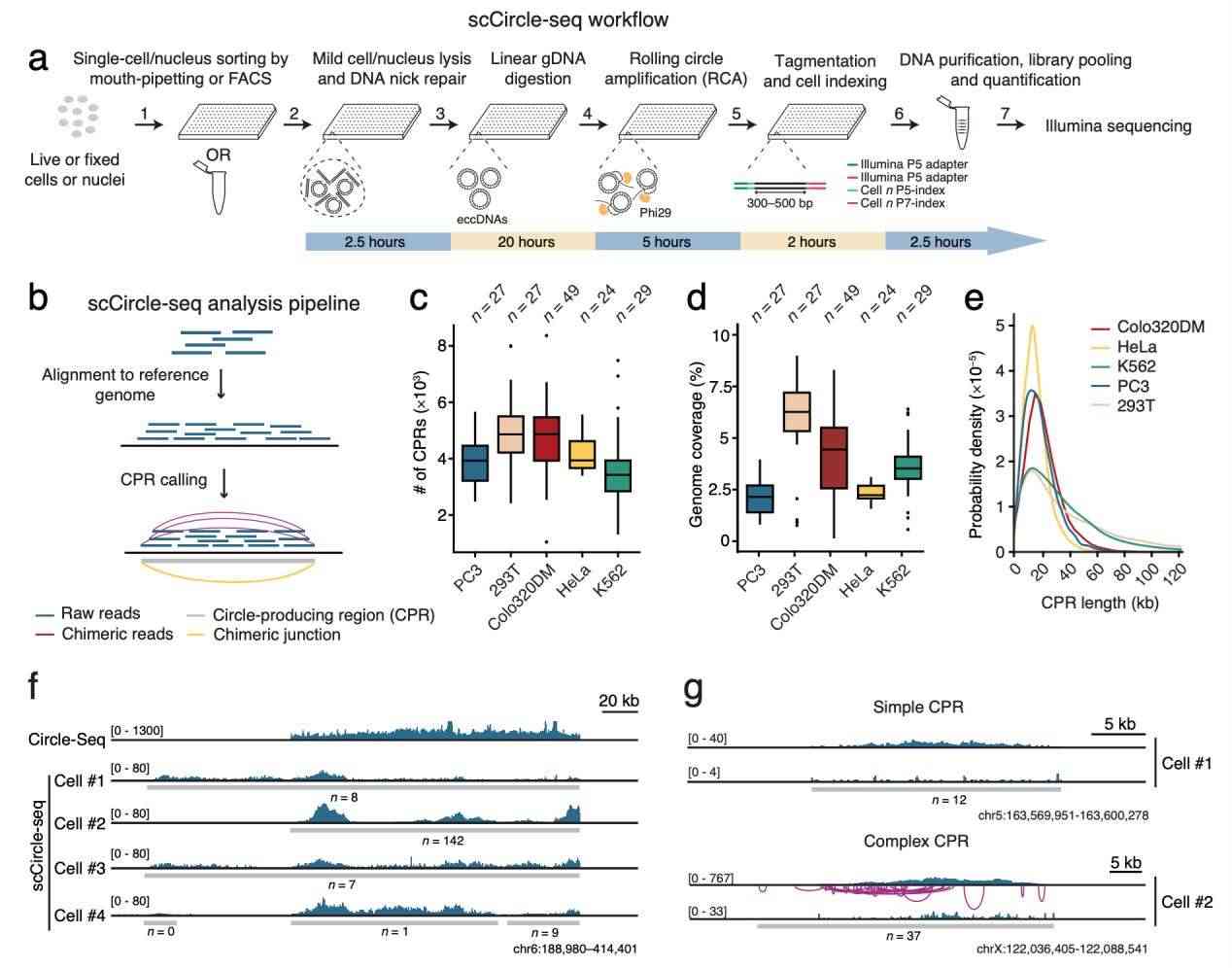 scCircle-seq workflow and data validation (Chen et al., 2024)
scCircle-seq workflow and data validation (Chen et al., 2024)
Genomic Atlas of eccDNA in Single Cells
The research team analyzed the genomic distribution of eccDNA detected by scCircle-seq to determine whether there is a specific pattern for eccDNA in single cells. They found that eccDNA exhibits substantial variability at the single-cell level but may reveal more defined patterns at the population level. Based on the frequency of corresponding CPRs (Circular DNA-Producing Regions) and their uniformity scores across all single cells within the same cell line, the team classified the eccDNA identified by scCircle-seq into four groups for further validation.
The results showed that the vast majority of detected eccDNA (88-99%) were categorized as Low Frequency, Low Uniformity (LFLU), while a small proportion was classified as High Frequency, High Uniformity (HFHU), predominantly observed in Colo320DM and 293T cells. These findings suggest that eccDNA is inherently highly heterogeneous, but scCircle-seq is capable of identifying different types of eccDNA.
Further exploration of the genomic distribution of the detected eccDNA was conducted by intersecting the identified CPRs with various genomic annotations. The analysis revealed that CPRs were enriched in chromatin regions marked by histone H3K9me3 (constitutive heterochromatin) and H3K27me3 (facultative heterochromatin), indicating that eccDNA formation is more frequent in heterochromatic regions.
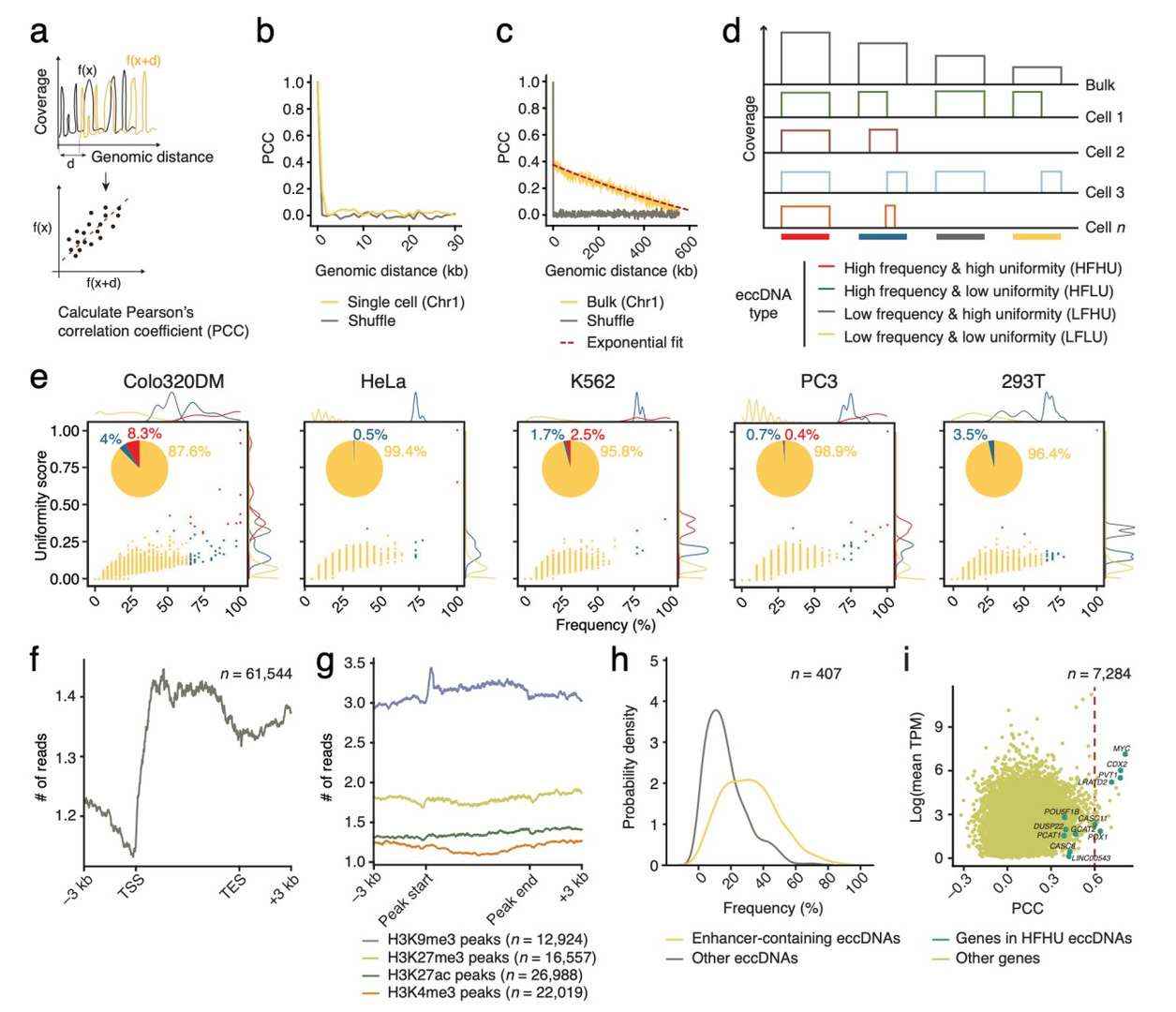 Genomic distribution of eccDNA identified by scCircle-seq (Chen et al., 2024)
Genomic distribution of eccDNA identified by scCircle-seq (Chen et al., 2024)
The research team next combined scCircle-seq with the full-length single-cell RNA sequencing method Smart-seq2 and applied this integrated approach to three cell lines (Colo320DM, HeLa, and PC3). They aimed to investigate the relationship between the eccDNA copy number detected by scCircle-seq and the gene expression levels associated with these eccDNAs.
The analysis revealed that there was generally a low correlation between gene expression levels and eccDNA copy numbers across the cells. However, in the case of large ecDNA containing known oncogenes (e.g., MYC), a positive correlation was observed between the copy number of eccDNA and the expression level of the same gene within single cells. This finding suggests that, in certain contexts, the presence of eccDNA may have a direct influence on the expression of genes located on these extrachromosomal elements, particularly oncogenes.
Cell Type Classification Based on eccDNA
The research team further investigated whether the eccDNA sequences within cells are random or if scCircle-seq could be used to identify specific eccDNA features associated with different cell types. To do so, they first represented the genomic distribution of CPRs detected by scCircle-seq as a cells × bins matrix, where cells refers to the number of single cells analyzed by scCircle-seq, and bins represents defined, continuous genomic windows of a certain length. They then used cisTopic, a computational framework, to cluster the cells and annotate each cluster with available genomic tracks.
The results demonstrated that this method successfully grouped cells of the same type together. Moreover, the analysis revealed that, while there is substantial variation in both the quantity and origin of eccDNA between cells of the same type, different cell types exhibit specific eccDNA features that partially reflect their epigenomic characteristics. This suggests that scCircle-seq could serve as a valuable tool for identifying cell-type-specific eccDNA profiles, offering insights into the epigenomic landscape of various cell types.
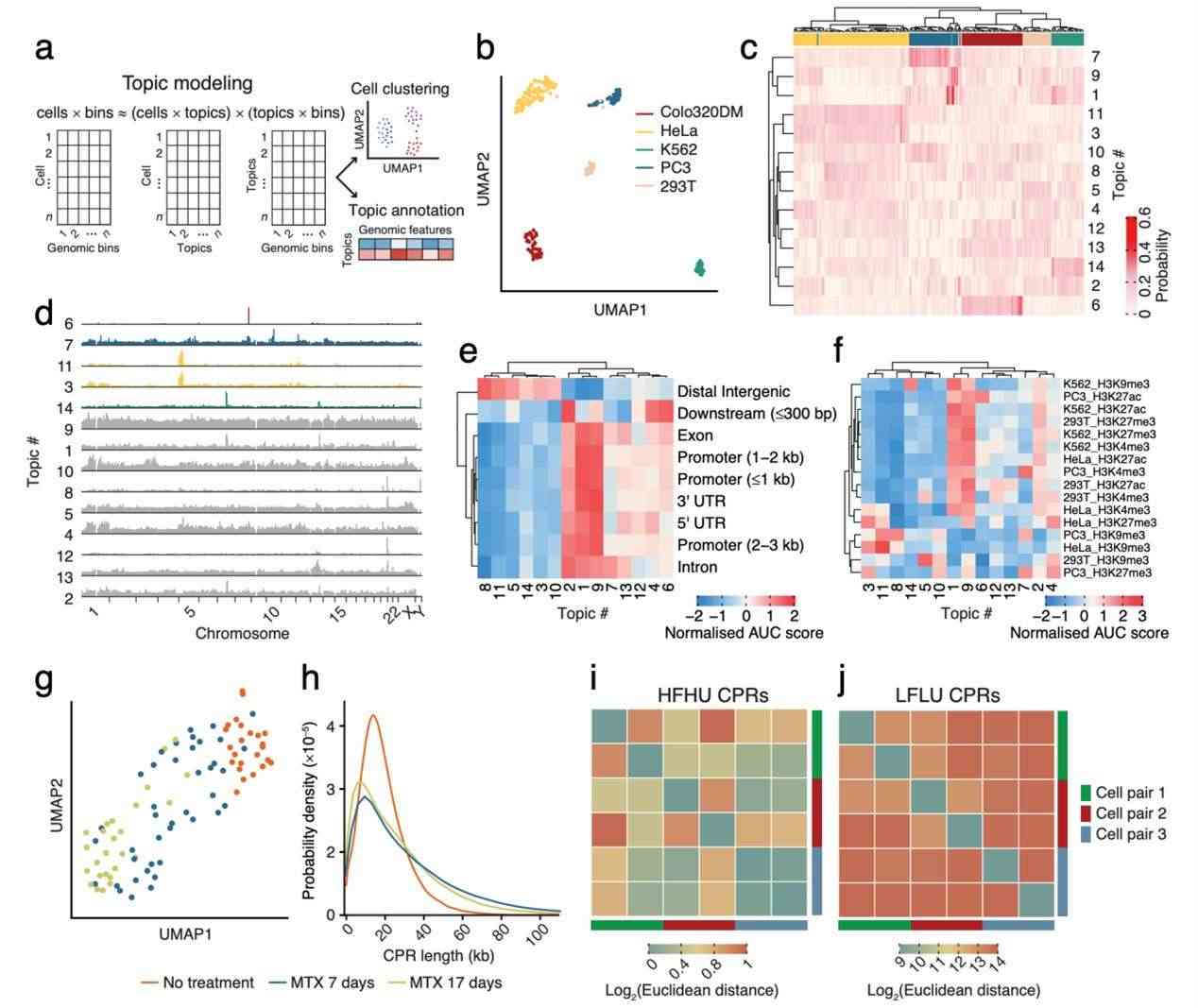 Cellular specificity of eccDNA (Chen et al., 2024)
Cellular specificity of eccDNA (Chen et al., 2024)
Application of scCircle-seq to Patient-Derived Tumor Samples
The research team applied scCircle-seq to nuclei extracted from three retrospectively collected frozen tumor samples: one from prostate adenocarcinoma (PRAD), one from triple-negative breast cancer (TNBC), and one from luminal B-type breast cancer (LumB), demonstrating the applicability of scCircle-seq to patient-derived tumor samples. A total of 55 PRAD nuclei, 87 TNBC nuclei, and 33 LumB nuclei were analyzed, yielding high-quality sequencing data. The ratio of circular DNA to linear DNA in all cells was close to 100%, consistent with results obtained from immortalized cell lines, indicating that scCircle-seq works well even with nuclei extracted from tumor biopsies.
The analysis revealed that PRAD cells exhibited the highest number of CPRs and the largest proportion of the genome covered by CPRs, followed by TNBC and LumB cells. This suggests that these tumor cells have distinct eccDNA profiles. Furthermore, dimensionality reduction (UMAP) and differential topic analysis of the single-cell CPRs genomic profiles showed that PRAD, TNBC, and LumB formed two separate clusters, indicating that these tumor types carry different eccDNA genomic landscapes, which aligns with their distinct tissue origins.
The team further cross-analyzed the identified CPRs from LumB and TNBC cells with somatic copy number alterations (SCNAs) identified in breast cancer from the The Cancer Genome Atlas (TCGA). The results revealed that amplification regions were significantly enriched in the CPRs of TNBC cluster 1 and LumB cells compared to TNBC cluster 2. Additionally, when analyzing the relationship between the number of identified eccDNAs and the copy number in specific genomic regions, the team found that the number of CPRs did not differ significantly in diploid or moderately amplified regions but was significantly higher in highly amplified regions.
These results indicate that the distribution of eccDNA in tumor cells reflects their SCNA status, with highly amplified genomic regions producing more eccDNA. This suggests that scCircle-seq can be a powerful tool for investigating the eccDNA landscape in tumor samples, offering insights into the relationship between eccDNA and genomic alterations in cancer.
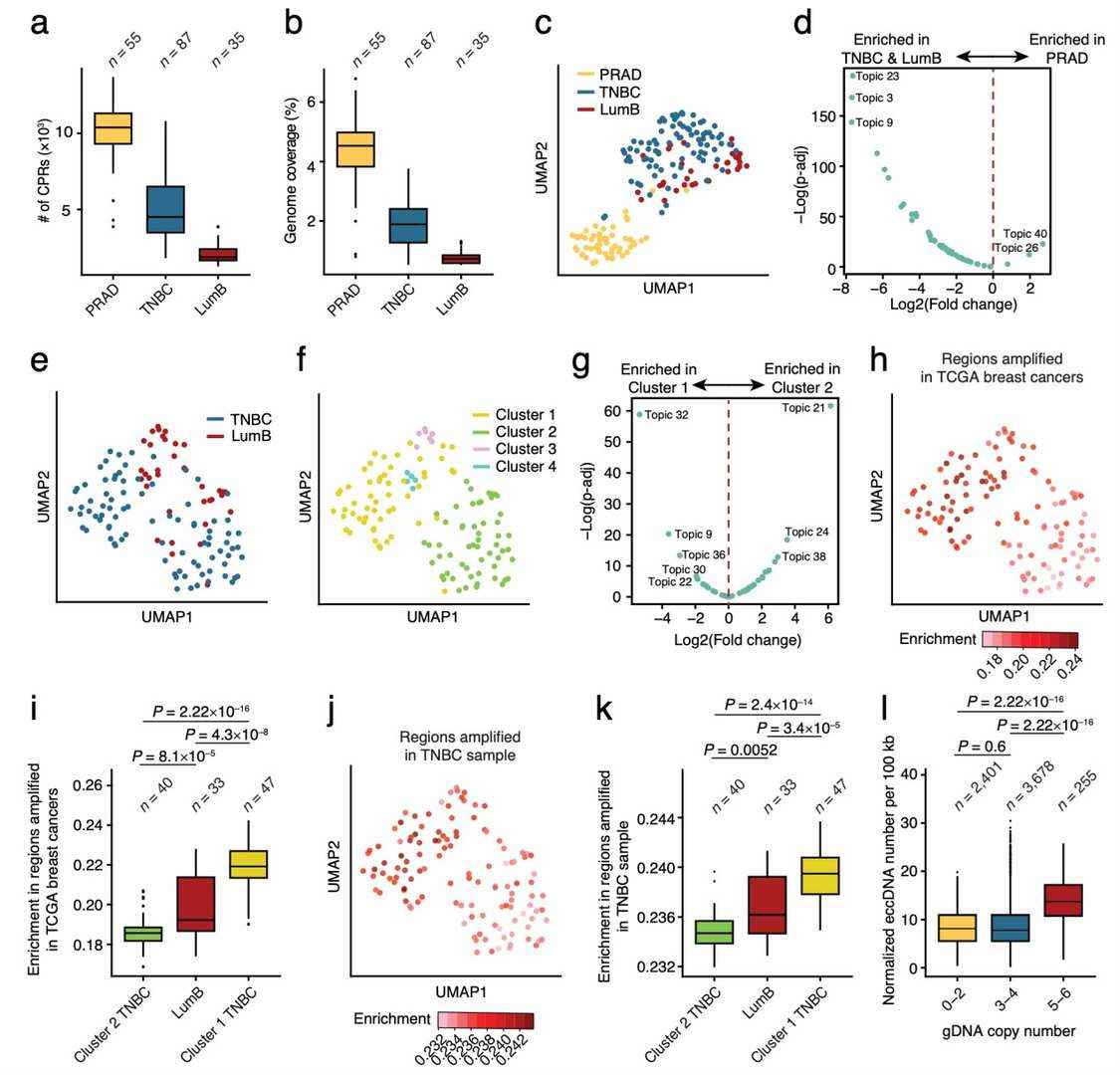 Application of scCircle-seq to patient-derived tumor samples (Chen et al., 2024)
Application of scCircle-seq to patient-derived tumor samples (Chen et al., 2024)
Take the Next Step: Explore Related Services
In conclusion, the scCircle-seq technique represents a groundbreaking advancement in the study of extrachromosomal circular DNA (eccDNA) at a single-cell level. By incorporating rolling circle amplification (RCA) and DNA nick repair, scCircle-seq offers enhanced sensitivity, enabling the detection of both abundant oncogene-containing eccDNA and rare eccDNA species. The method's streamlined workflow minimizes the chances of overlooking low-abundance eccDNA, which significantly improves the overall comprehensiveness of eccDNA analysis.
What sets scCircle-seq apart is its adaptability across various cell types and tissues, which allows for in-depth exploration of eccDNA's role in diverse biological contexts. This versatility paves the way for novel insights into the functional contributions of eccDNA in both healthy and diseased states, particularly in cancer. EccDNA has been increasingly recognized for its involvement in gene expression regulation, tumor progression, and the development of drug resistance, with scCircle-seq poised to reveal new facets of these mechanisms.
Moreover, scCircle-seq's simplicity and scalability make it an ideal tool for large-scale studies, particularly those involving patient-derived tumor samples. By identifying specific eccDNA markers, this method could potentially offer new avenues for early cancer diagnosis, monitoring progression, and assessing therapeutic responses, thereby holding promise for significant improvements in cancer diagnostics.
In essence, scCircle-seq provides a versatile and expandable framework for unraveling the complexities of eccDNA across various biological systems. As it continues to evolve, the method offers immense potential for clinical applications, ultimately contributing to more effective cancer research and personalized therapeutic strategies.
Reference

CD Genomics is transforming biomedical potential into precision insights through seamless sequencing and advanced bioinformatics.
We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy