What Is SNP Microarray
SNP microarray, a specialized type of microarray technology, is geared towards the detection of Single Nucleotide Polymorphisms (SNPs), characterizing variations at a single base pair in the genome. Emerged in recent years, SNP microarray technology represents a high-throughput, large-scale genetic testing platform. The working principle largely entails the hybridization of fragmented single-stranded DNA with an array comprising hundreds of thousands of unique nucleotide probe sequences.
Single nucleotide polymorphism (SNP) refers to the difference of single nucleotide at the same position in the genomic DNA sequence. In general, a single nucleotide variation with a frequency greater than 1% is called an SNP. The SNPs involve only a single base variation, which can be caused by a single base transition or transversion. There is approximately one SNP per 1000 bases in the human genome, and the total number of SNPs in the human genome is around 3 x 106.
SNP has become the third generation of genetic markers. The characteristics of SNPs make them more suitable for studies on genetic pleiotropy in complex traits and diseases, and population-based gene recognition than other polymorphic markers. SNPs have significant importance in biomedical research. SNPs have been used in genome-wide association studies as high-resolution markers in gene mapping related to diseases or normal traits. SNPs without an observable impact on the phenotype (called silent mutations) may also be useful due to their quantity and stable inheritance over generations.
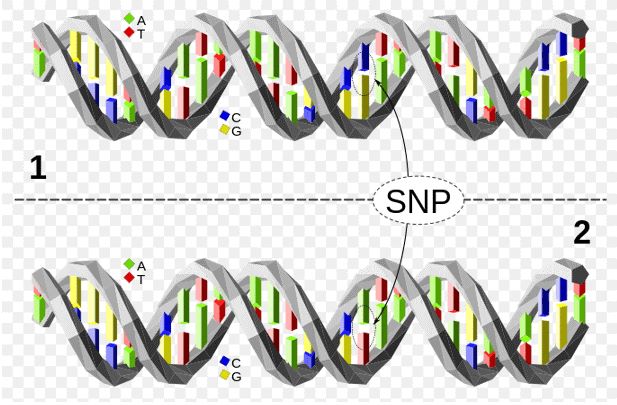 Figure 1. The upper DNA molecule differs from the lower DNA molecule at a single base-pair location (a C/A polymorphism).
Figure 1. The upper DNA molecule differs from the lower DNA molecule at a single base-pair location (a C/A polymorphism).
The DNA microarray refers to a gene chip with a large number of probe DNA sequences in a specific arrangement immobilized on a solid substrate. The principle is the nucleic acid hybridization theory, and the detected sample DNA is hybridized with the DNA microarray and extended. Subsequently, the fragments of the non-complementary binding reaction on the chip are washed away, and the gene chip is subjected to laser confocal scanning. Fluorescence signal intensities are then measured and interpreted as the abundance of different genes through certain data processing.
Service you may intersted in
Principles of SNP Microarray
The fundamental principle of SNP microarrays involves a single hybridization strategy where DNA samples, for scrutiny, undergo hybridization with probes integrated on the chip. The copy number at each locus can subsequently be determined by comparing signal intensities across various samples.
SNP microarrays take advantage of the differences in SNP loci between individuals. Conventionally, the detection of SNP sites is based on DNA sequence alignment. Within SNP microarrays, probes featuring different SNP variations are employed. These probes specifically match with the SNP sites. Through hybridization of DNA from the samples, the genotype of each SNP site can be ascertained. Further, the frequency of different genotypes can be determined by gauging the intensity of the signal.
Compared to traditional single-cell diagnosis methods, SNP microarray serves as a high-throughput approach, capable of conducting thousands of reactions on the oligonucleotide chip surface simultaneously.
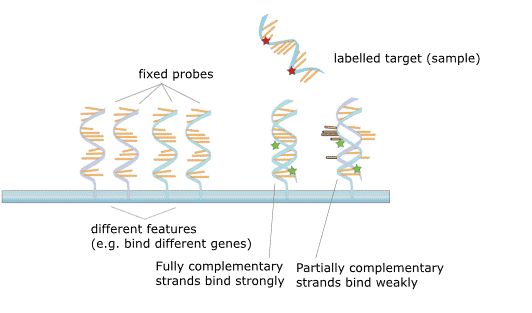 Figure 2. The principle of microarray.
Figure 2. The principle of microarray.
Workflow of SNP Microarray
The general workflow of SNP microarray is shown in figure 3. Briefly speaking, there are several processes: SNP chip fabrication, sample genomic DNA preparation, hybridization, and fluorescence scanning.
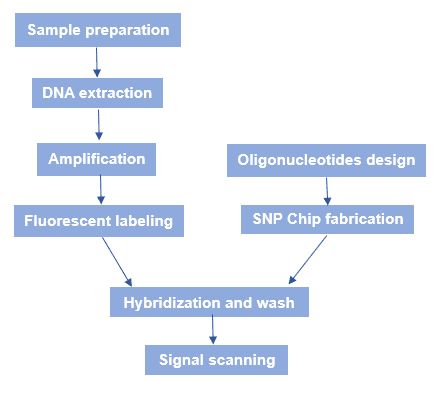 Figure 3. The workflow of SNP microarray.
Figure 3. The workflow of SNP microarray.
Probe Design
The process involves the collection of genomic sequence information from the target Single Nucleotide Polymorphism (SNP) locales. Known SNP information is then aligned with the reference genome sequence, which determines the position and variable basis of SNP sites. Utilizing sequence information around the SNP sites, a set of specific probes are designed. This guarantees that probes will selectively pair with the variable bases at the target SNP sites.
Upon designing the probes, in vitro experiments are conducted to evaluate the hybridization performance of probes, including hybridization efficiency under conditions such as temperature and salt concentration. The length of the probes is controlled, typically ranging between 20 to 70 bases in order to ensure stable hybridization and reliable signal detection. In the probe design stage, the reverse complement sequence of the target SNP location is also taken into account to facilitate bidirectional mutation detection.
SNP chip fabrication
The predesigned oligonucleotide or cDNA chips are arranged orderly and in high density on the glass carrier to make microarrays or core chips. The fabrication methods of the chip include light-guided in-situ synthesis, chemical spray method, contact dot coating method, in-situ DNA controlled synthesis, non-contact micromechanical printing method TOPSPOT® and soft lithography reproduction, etc. A total of 4x105 different DNA molecules can now be placed on a 1 cm2 chip. Affymetrix GeneChip® technology allows high-density, in situ oligonucleotide synthesis, and has been a pioneer in this field.
Sample genomic DNA preparation
In genomic DNA sample preparation, an initial step is to acquire suitable sample material. Following this, it's imperative to extract genomic DNA and then amplify it via Polymerase Chain Reaction (PCR). Lastly, by employing different fluorescent dyes, the samples are labeled for further analysis.
The concentration and purity of the DNA samples are of paramount importance. These can be gauged using spectrophotometric techniques, such as colorimetry, ultraviolet absorption spectroscopy, or assays utilizing fluorescent dyes. Such procedures ensure that the extracted DNA is of sufficient quality and does not possess any evident contamination. Furthermore, aligning with the experimental requirements, an appropriate labeling method is chosen. Commonly implemented labeling techniques encompass fluorescence labeling, biotin labeling, as well as radioactive isotope labeling.
Hybridization and scan
Moving onto hybridization and scanning, the fluorescently labeled genomic DNA hybridizes with the SNP microarray under suitable reaction conditions. Post-washing, the fluorescence is scanned with a specifically designed fluorescence scanner. Thereafter, the data is processed through computer image analysis and subsequently submitted for bioinformatics analysis of the target genes.
Foremost, the extent of optimization of various hybridization conditions — such as temperature, salt concentration, and hybridization time — are of paramount importance to ensure sufficient binding of DNA with the probes. Subsequently, during elution, the elution conditions should be carefully controlled to ensure only DNA that binds with the probe is retained on the chip, consequently averting the possibility of unbound DNA compromising the accuracy of the results.
Advantages and Limitations of SNP Microarray
Advantages of this process include the ability to concurrently detect multiple SNP sites rapidly- yielding a large amount of SNP data in a single experiment. This procedure is applicable in a broad range of fields, such as genome-related research, individual genotype analysis, and disease susceptibility studies and it provides a higher level of accuracy in detecting copy number variation than next-generation sequencing, including whole-genome sequencing (WGS). It also possesses a higher resolution than array comparative genomic hybridization (aCGH), capable of detecting heterozygote neutral loss of copy number, certain forms of uniparental disomy, and changes in ploidy.
However, limitations exist as this method is constrained by known SNP information and cannot detect unknown SNP mutations. Labeling methods may introduce bias and affect the accuracy of results. Moreover, handling extensive data requires complex analysis methods and computation, and the cost for chip preparation, reagents, and equipment is relatively high.
Application of SNP Microarray
SNP microarrays have become a frequently employed instrument in various domains, including genomic research, personalized medicine, disease diagnosis, and drug development.
SNP genotyping, the process of identifying the genetic variants of an individual, allows us to determine a person's genetic characteristics, including different alleles at various loci throughout the genome. Furthermore, by scrutinizing the frequency distribution of SNP loci across different populations or ethnic groups, we can gain insights into the genetic structure and evolutionary history of these groups.
Comparative analysis of SNP microarray data between patients with a disease and healthy control participants can identify SNP loci associated with genetic disorders, thereby providing a window into the genetic underpinnings of the disease. The SNP technique presents us with opportunities to discover correlations with complex diseases (such as cancer, cardiovascular disease, mental disorders, etc.) and explore their genetic risk factors.
An analysis of the patient's genetic profile offers a means to predict their susceptibility to environmental influences and disease predispositions, thereby providing a basis for personalized disease prevention and health management. It is used to detect genomic structural variations, such as SNPs anCNVs, revealing the structure and function of the genome.
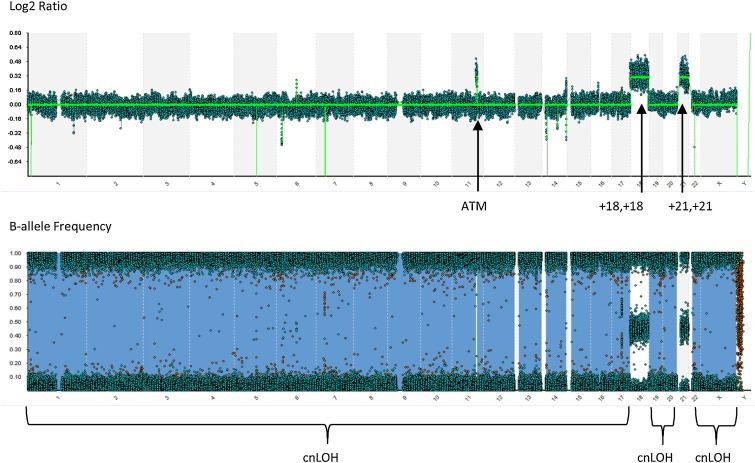 Figure 4. Whole genome view from a SNP-microarray performed on the diagnostic bone marrow specimen submitted for investigation of ALL. (Berry et al., 2019)
Figure 4. Whole genome view from a SNP-microarray performed on the diagnostic bone marrow specimen submitted for investigation of ALL. (Berry et al., 2019)
Additionally, SNP microarrays are widely employed in genome-wide association studies (GWAS). By analyzing SNPs across large samples, researchers can identify SNP loci related to specific phenotypes or diseases and explore their genetic foundation.
In recent years, methods underpinned by SNP microarrays have been incorporated into prenatal diagnostics. This is primarily due to the capability of SNP analysis to diagnose regions of homozygosity (ROH), unbalanced translocations, and other aspects in diagnosing genomic diseases. These techniques boast considerable advantages over the traditional chromosomal karyotype analysis, marking them as indispensable first-line genetic diagnostic technologies within contemporary clinical settings.
Difference Between CGH and SNP microarray
The technologies of CGH microarrays and SNP microarrays serve vital roles in genome analysis. However, differences exist in their principles, applications, merits, and demerits.
Principles:
CGH Microarray: Deployed for detecting Copy Number Variations (CNV) within genomic DNA, the principle of CGH involves labeling the sample DNA and reference DNA concurrently, followed by their hybridization onto the chip. Copy-number variations in the sample DNA can be determined by contrasting the signal intensity between the sample DNA and the reference DNA.
SNP Microarray: The SNP microarray is designed primarily for identifying Single Nucleotide Polymorphisms, i.e., variations at single base pairs in the genome. Its underlying principle lies in hybridizing the sample DNA with SNP probes on the chip and determining genotypes at each SNP locus through the signal strength of the hybridization between the probe and sample DNA.
Applications:
CGH Microarray: Its major application is detecting CNVs, such as gene duplication, deletion, or amplification, and identifying chromosomal structural variations. It is widely used in cancer genomics research and genetic disorder diagnosis.
SNP Microarray: Predominantly utilized for detecting Single Nucleotide Polymorphisms, including genotype identification, individual genomic analysis, susceptibility research, population genetic structure analysis, etc. It has extensive utility in Genome-Wide Association Studies (GWAS).
Advantages and Disadvantages:
CGH Microarray: Advantages: It allows concurrent detection of large CNV segments in the genome with a wide coverage, which is suitable for discovering novel CNVs. Disadvantages: It does not offer detailed SNP data, thus incapable of detecting single base pair variations, demonstrating a relatively weak capacity for small fragment variation detection.
SNP Microarray: Advantages: It provides direct SNP data, making it suitable for individual genotyping and disease-related studies, boasting high resolution and specificity. Disadvantages: It can only detect known SNP locations, providing no information on unknown variations and direct CNV detection.
At CD Genomics, we are dedicated to providing reliable SNP genotyping services, including genotyping by sequencing, SNP microarray, MassARRAY SNP genotyping, SNaPshot Multiplex System for SNP genotyping, and TaqMan SNP genotyping.
References:
- Iwamoto K, Bundo M, Ueda J, et al. Detection of chromosomal structural alterations in single cells by SNP arrays: a systematic survey of amplification bias and optimized workflow. Plos One, 2007, 2(12): e1306.
- Ho C C, Mun K S, Naidu R. SNP array technology: an array of hope in breast cancer research. Malaysian Journal of Pathology, 2013, 35(1):33-43.
- Ahmad A, Iqbal M A. Significance of genome-wide analysis of copy number alterations and UPD in myelodysplastic syndromes using combined CGH-SNP arrays. Current Medicinal Chemistry, 2012, 19(22).
- Sund K L, Zimmerman S L, Thomas C, et al. Regions of homozygosity identified by SNP microarray analysis aid in the diagnosis of autosomal recessive disease and incidentally detect parental blood relationships. Genetics in Medicine Official Journal of the American College of Medical Genetics, 2013, 15(1):70-78.
- Liu S, Zhou Z, Lu J, et al. Generation of genome-scale gene-associated SNPs in catfish for the construction of a high-density SNP array. Bmc Genomics, 2011, 12(1):53.
- Fang Z, Che D, Gu X. LncRNA HULC polymorphism is associated with recurrent spontaneous abortion susceptibility in the Southern Chinese Population. Frontiers in Genetics, 2019, 10: 471328.
- Berry N K, Scott R J, Rowlings P, et al. Clinical use of SNP-microarrays for the detection of genome-wide changes in haematological malignancies. Critical reviews in oncology hematology, 2019, 142: 58-67.


 Sample Submission Guidelines
Sample Submission Guidelines
 Figure 1. The upper DNA molecule differs from the lower DNA molecule at a single base-pair location (a C/A polymorphism).
Figure 1. The upper DNA molecule differs from the lower DNA molecule at a single base-pair location (a C/A polymorphism). Figure 2. The principle of microarray.
Figure 2. The principle of microarray. Figure 3. The workflow of SNP microarray.
Figure 3. The workflow of SNP microarray. Figure 4. Whole genome view from a SNP-microarray performed on the diagnostic bone marrow specimen submitted for investigation of ALL. (Berry et al., 2019)
Figure 4. Whole genome view from a SNP-microarray performed on the diagnostic bone marrow specimen submitted for investigation of ALL. (Berry et al., 2019)