CD Genomics has been providing the accurate and affordable genotyping by sequencing services for couple of years. We employ the latest Illumina sequencing platforms to obtain genetic variations information in a more efficient way.
The Introduction of Genotyping by Sequencing (GBS)
Genotyping by Sequencing (GBS) represents a groundbreaking technique in genomic research, facilitating high-throughput detection of single nucleotide polymorphisms (SNPs) across extensive populations. As a methodology within the Reduced-Representation Genome Sequencing (RRGS) family, GBS utilizes restriction enzyme digestion to reduce genome complexity, followed by high-throughput sequencing to expose genetic variants. This technique is particularly advantageous for organisms with incomplete genomic resources, as it obviates the need for prior genome sequence knowledge.
GBS functions by employing restriction enzymes to cleave genomic DNA at specific loci, which are subsequently ligated with DNA-barcoded adapters. The generated fragments are then sequenced, offering a comprehensive representation of SNPs across the genome. This method is distinguished by its capability to provide extensive SNP coverage in gene-rich regions, making it indispensable for precise genetic analysis and breeding initiatives.
Difference between RAD Sequencing and GBS
- GBS utilizes restriction enzymes to create a reduced-complexity library, which is then sequenced. This approach is streamlined, involving fewer preparatory steps than RAD-seq and requiring a lesser quantity of DNA. Notably, GBS does not necessitate a size selection step, making it an efficient method for SNP discovery across a diverse array of species. It is particularly advantageous for its capability to deliver high coverage and cost-effective genotyping on a large scale.
- Conversely, RAD-seq also employs restriction enzymes to generate DNA fragments for sequencing. However, RAD-seq involves a more intricate library preparation process, including a size selection step. This additional step aims to capture and sequence specific genomic regions around the restriction sites, thus providing a targeted approach to studying genetic variation in designated areas of the genome. The inclusion of these extra steps allows RAD-seq to focus on particular loci, thereby offering detailed insights into genetic variation within specific genomic contexts.
Advantages of Our GBS Services
- Cost-Effectiveness and Scalability: GBS represents a cost-effective and scalable approach to genotypic analysis. By reducing sequencing expenditures, GBS facilitates the inclusion of large sample sizes, thereby rendering it exceptionally suited for extensive genomic projects. This economic efficiency, coupled with its scalability, makes GBS a favorable option for both small-scale and large-scale research endeavors.
- Comprehensive Workflow: Our GBS services encompass an integrated workflow, providing a seamless journey from DNA extraction through to variant calling. This comprehensive service ensures accuracy and efficiency at every stage of the process, delivering reliable and precise genetic data. By overseeing the entire workflow, we guarantee high-quality outcomes and a streamlined experience for researchers.
- High Throughput and Rapid Turnaround: Leveraging advanced sequencing technologies, our GBS platform can process thousands of samples simultaneously. This capacity for high throughput translates to rapid turnaround times, allowing for the timely acquisition of genotypic data. Researchers can expect quick and dependable results, facilitating swift progression in their genomic studies.
- High-Quality Genotyping: Our GBS services attain an impressive genotyping rate exceeding 95% per locus. This high level of reliability ensures the acquisition of robust and high-quality genetic data, critical for accurate genetic analysis and subsequent applications in breeding and research.
Application of GBS
- Genome-Wide Association Studies (GWAS): GBS has emerged as an invaluable tool in the realm of GWAS, which aim to pinpoint genetic variants linked to complex traits and diseases. By delivering a comprehensive catalog of SNPs spread across the entire genome, GBS facilitates the identification of genetic markers tied to specific phenotypic traits.
- Genetic Diversity and Linkage Analysis: GBS also plays a critical role in studies focused on genetic diversity, aiding researchers in evaluating the genetic variation both within and between populations. Such assessments are pivotal for understanding the intricate patterns of genetic diversity and are instrumental in guiding decisions within breeding programs. Furthermore, GBS enhances genetic linkage analysis by mapping genetic loci associated with particular traits, thereby assisting in the identification of genes responsible for these traits.
- Molecular Marker Discovery and Genomic Selection: The utility of GBS extends to the realm of molecular marker discovery, providing a robust platform for identifying new genetic markers that can be employed in a variety of applications, including breeding and genetic research. This technique is particularly advantageous for species with incomplete genomic data, as it does not require a reference genome. Additionally, GBS supports the principles of genomic selection by enabling researchers to make informed choices about desirable traits, grounded in comprehensive genetic data.
Genotyping by Sequencing (GBS) Workflow
The workflow of GBS at CD Genomics involves several key steps:

Service Specifications
Sample Requirements
|
|
Click |
Sequencing Strategy
|
| Bioinformatics Analysis
We provide multiple customized bioinformatics analyses:
|
Analysis Pipeline
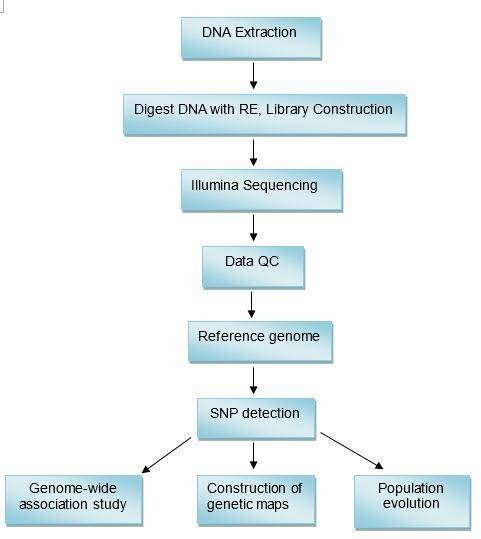
Deliverables
- The original sequencing data
- Experimental results
- Data analysis report
- Details in GBS for your writing (customization)
CD Genomics is proud to provide genotyping by sequencing service to advance your population based genotyping studies in an effective and economic way. Please contact us for more information and a detailed quote. CD Genomics is the licensed service provider for this technology. Keygene N.V. owns patents and patent applications protecting its Sequence Based Genotyping technologies.
Partial results are shown below:
1. What is the definition of a GBS Tag?
A GBS Tag refers to a sequence of reads adjacent to a restriction enzyme cut site. The genomic coverage captured by GBS is determined by multiplying the number of Tags by the length of a single read. For instance, utilizing HiSeq 4000 PE150 sequencing, the genomic coverage can be calculated as follows:
GBS captured genomic range=100,000 Tag x 150bp/Tag=100000X150=15 M
If the average sequencing depth per sample is 10x per Tag, the sequencing data volume per sample would be:
15Mx 10=150 M
2. How to select the number of Tags?
The required number of Tags varies depending on the specific research objectives. For example, GWAS might necessitate tens of thousands of high-density molecular markers, whereas studies on phylogenetic relationships or linkage analysis may only require a few hundred to a few thousand molecular markers to achieve satisfactory results. Hence, it is crucial to first evaluate the necessary number of Tags based on the study's requirements and then select an appropriate number of Tags accordingly.
For species with a genome size less than 1 Gb undergoing genetic linkage map studies, a common recommendation is to utilize approximately 100,000 Tags. Adjustments to the number of Tags can be made based on specific research needs.
Table 1. Commonly used GBS Tag numbers (e.g., for genetic linkage maps).
| Genome Size | Number of Tags |
|---|---|
| Below 10G | ≥10W tag |
| 1-2G | ≥15W tag |
| 2-3G | ≥20W tag |
3. Can GBS be used for non-reference species?
GBS can indeed be utilized for non-reference species to obtain SNP markers. However, the lack of annotated genomic information in non-reference species presents a significant challenge, often rendering the identification of candidate genes unfeasible. For tasks such as quantitative trait loci (QTL) mapping, association studies, or the mining of genes related to domestication traits, it is advisable to utilize reference species to achieve more accurate and informative results.
4. Can GBS be applied to polyploid species?
GBS technology is applicable to polyploid species. A salient example is the successful application of GBS to hexaploid oat species for genetic mapping in 2014. Polyploid species are characterized by their complex ploidy levels, which can include both autopolyploids and allopolyploids, as well as tetraploids and hexaploids. Each scenario requires specific analytical considerations. Current research efforts have already begun to employ GBS for genetic mapping in polyploid crops such as wheat and cotton.
5. Is GBS suitable for inter-species research?
GBS leverages restriction enzymes for genome capture, thereby facilitating the development of SNP markers needed for genetic linkage and population genetic analyses. Significant genetic divergence between samples can result in non-uniform capture of restriction fragments across samples and a paucity of shared SNPs. Consequently, GBS is predominantly suited for studies at the intra-species level. Nonetheless, in rare cases where there is a close phylogenetic relationship between different species within the same genus and minimal genetic divergence, GBS can be employed effectively for phylogenetic studies.
Genetic differentiation of grain, fodder and pod vegetable type cowpeas (Vigna unguiculata L.) identified through single nucleotide polymorphisms from genotyping-by-sequencing
Journal: Molecular Horticulture
Impact factor: 10.6
Published: 28 March 2022
Background
Cowpea (Vigna unguiculata) is an important legume used globally for various purposes, including fresh vegetables, dry grains, and fodder. It comes in three main subspecies: ssp. sesquipedalis (vegetable types), ssp. unguiculata (pulse types), and ssp. cylindrica (fodder types). Originating in Africa, cowpea spread to South Asia and the Americas through trade. Recent advancements using Genotyping-by-Sequencing (GBS) have improved SNP discovery and genetic diversity analysis, providing valuable insights for breeding and genetic studies.
Materials & Methods
Sample Preparation
- Cowpea
- DNA extraction
Sequencing
- GBS
- Illumina HiSeq 2000 platform
- Single-end sequencing
- SNP calling
- Population structure analyses
- phylogeny analyses
Results
1. SNP Discovery and Distribution
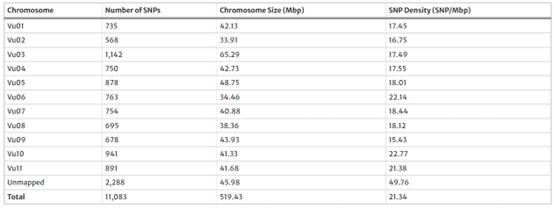
Out of 63,947 identified polymorphisms, 11,083 high-quality SNPs were retained, showing an average density of one SNP every 21.3 Mbp across the genome. SNPs were distributed unevenly, with higher density near sub-telomeric regions.
Table 1 Number of high-quality single nucleotide polymorphism (SNP) loci found by genotyping-by-sequencing
2. Principal Component Analysis (PCA)
PCA revealed clear differentiation between yardlong beans, grain cowpeas, and fodder cowpeas, with distinct clustering for each type and intermediate clustering for fodder types.
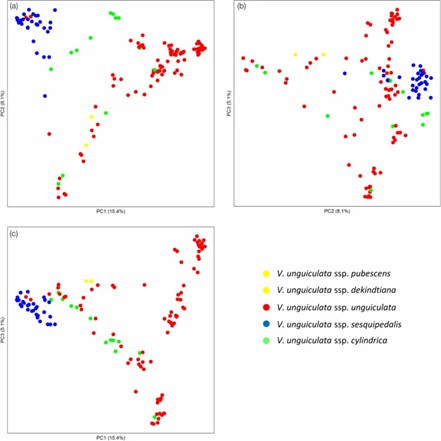 Fig 1. Visualization of the genetic relationships among three domesticated cowpea subspecies by Principal Component Analyses.
Fig 1. Visualization of the genetic relationships among three domesticated cowpea subspecies by Principal Component Analyses.
3. Population Structure
Bayesian clustering identified two major groups at K=2, and four sub-groups at K=4, reflecting taxonomic and growth type differences, with evidence of genetic admixture among cowpea subspecies.
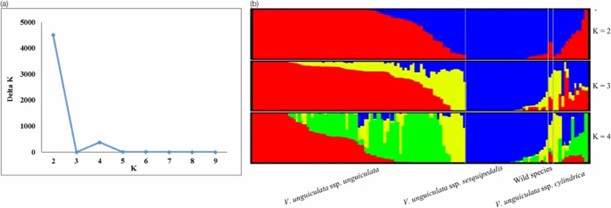 Fig 2. Population structure analysis for 130 cowpea accessions.
Fig 2. Population structure analysis for 130 cowpea accessions.
4. Phylogenetic Analysis
Phylogenetic tree identified three main clusters: vegetable cowpeas, grain cowpeas, and fodder cowpeas, with some grain types clustering with vegetable types, indicating possible genetic introgression.
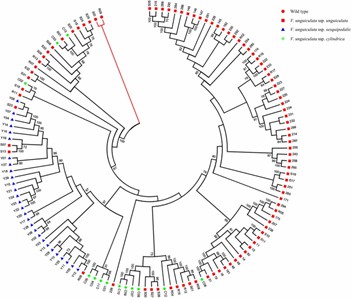 Fig 3. Rooted phylogenetic tree of 130 accessions from three cowpea subspecies and two wild relative species. Bootstrap values > 50% are marked above nodes.
Fig 3. Rooted phylogenetic tree of 130 accessions from three cowpea subspecies and two wild relative species. Bootstrap values > 50% are marked above nodes.
5. Genetic Diversity and Divergence
High genetic divergence was observed between grain and vegetable cowpeas, with notable outlier regions linked to traits like pod length and seed coat pattern. Divergence was concentrated at chromosome ends, indicating selective pressure and genomic diversity.
Conclusion
This study used a large SNP dataset to reveal genetic differences among three cowpea subspecies. It found that ssp. unguiculata and ssp. cylindrica are closely related, while ssp. sesquipedalis is distinct. Key selection signatures were identified on chromosomes Vu03 and Vu08, related to traits like pod length and flower scent. The findings offer insights into cowpea domestication and provide a basis for future breeding and gene mapping.
Reference
- Wu X, Cortés A J, Blair M W. Genetic differentiation of grain, fodder and pod vegetable type cowpeas (Vigna unguiculata L.) identified through single nucleotide polymorphisms from genotyping-by-sequencing. Molecular Horticulture, 2022, 2(1): 8.
Here are some publications that have been successfully published using our services or other related services:
Use of biostimulants for water stress mitigation in two durum wheat (Triticum durum Desf.) genotypes with different drought tolerance
Journal: Plant Stress
Year: 2024
The Restriction-Modification Systems of Clostridium carboxidivorans P7
Journal: Microorganisms
Year: 2023
In the land of the blind: Exceptional subterranean speciation of cryptic troglobitic spiders of the genus Tegenaria (Araneae: Agelenidae) in Israel
Journal: Molecular Phylogenetics and Evolution
Year: 2023
Genetic Modifiers of Oral Nicotine Consumption in Chrna5 Null Mutant Mice
Journal: Front. Psychiatry
Year: 2021
A high-density genetic linkage map and QTL identification for growth traits in dusky kob (Argyrosomus japonicus)
Journal: Aquaculture
Year: 2024
Genomic and chemical evidence for local adaptation in resistance to different herbivores in Datura stramonium
Journal: Evolution
Year: 2020
See more articles published by our clients.


 Sample Submission Guidelines
Sample Submission Guidelines
