CD Genomics offers custom Taqman assays to perform SNP genotyping, enabling the identification and amplification of polymorphisms.
The Introduction of TaqMan SNP Genotyping

TaqMan is a commonly used SNP genotyping method developed by Life Technologies. It is an advanced, mature, validated, and widely used technology. The TaqMan SNP genotyping assay is also called the "50-nuclease allelic discrimination assay". Each TaqMan genotyping assay contains two primers for amplifying the sequence of interest and two allele-specific and differently labeled TaqMan minor groove binder (MGB) probes for allele detection. Each allele-specific MGB probe is labeled with a fluorescent reporter dye (either a FAM or a VIC reporter molecule) in the 5' end and is attached with a fluorescence quencher to the 3' end. While the probe is intact, the proximity of the quencher dye to the reporter dye suppresses the reporter fluorescence. During the PCR amplification step, if the allele-specific probe is perfectly complementary to the SNP allele, it will hybridize to the target DNA segment and then get degraded by the 5'-nuclease activity of the Taq polymerase. The degradation of the probe results in the separation of the fluorophore from the quencher molecule, generating a detectable fluorescent signal. If a single point mismatch is present between the probe and the target DNA strand due to a SNP, the binding of the probe to DNA is destabilized during strand displacement in PCR, which will reduce the efficiency of probe cleavage and quenching of the fluorescent reporter dye.
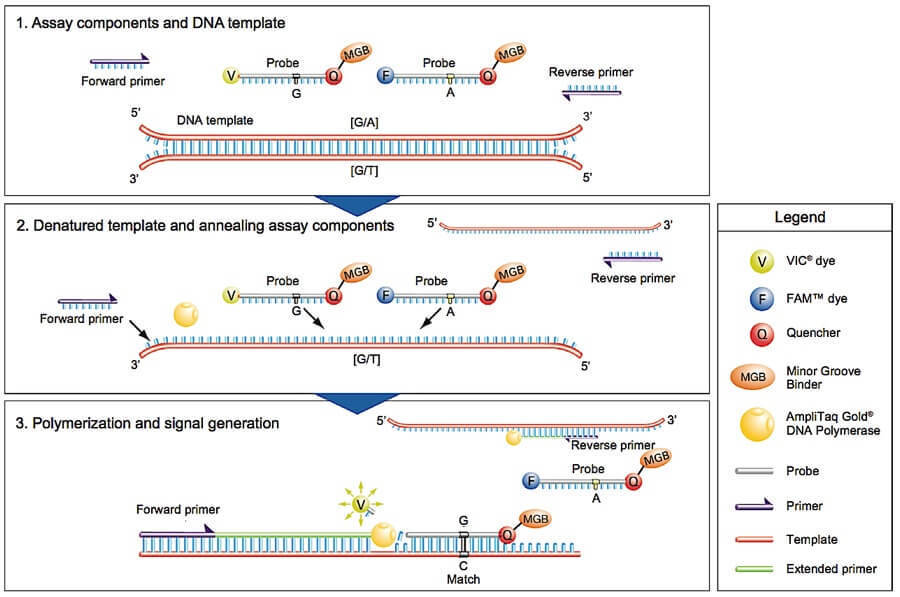
The assay can be performed using an ABI Prism 7900HT sequence detection system at high throughput and with low running costs. The ABI Prism 7900HT system can distinguish and quantify the two colors (FAM or VIC), and genotypes can be generated for many samples in a few minutes.
Advantages of TaqMan SNP Genotyping
- Simple and straightforward: TaqMan SNP genotyping involves simple mixing of samples and reaction components in a closed system, requiring no complex procedures and being easy to implement.
- Real-time detection: By detecting fluorescence changes during the PCR reaction, TaqMan SNP genotyping enables real-time monitoring of SNP genotypes, providing rapid results.
- Avoidance of post-PCR handling: With no need for post-PCR procedures, it eliminates labor costs and the risk of cross-contamination associated with post-PCR handling, reducing experimental errors.
- Low sample requirement: Due to the sensitivity of PCR reactions, TaqMan SNP genotyping requires minimal sample quantities, making it suitable for analysis of limited samples.
- High-throughput sample detection: With the advancement of fluorescence quantitative PCR instruments, large batches of samples can be simultaneously detected in a single reaction plate, improving detection efficiency and throughput.
- High sensitivity and specificity: TaqMan SNP genotyping offers high sensitivity and specificity, accurately identifying SNPs and avoiding misjudgments and confusion.
- Low sample input requirement with ~10ng/genotype
- Reasonable for large sample size
- Customized project design
- Knowledgeable scientists with considerable PCR experience
Application of TaqMan SNP Genotyping
- Especially advantageous for SNP analysis scenarios with a relatively modest number of loci, yet with sample sizes exceeding 1500 cases, as larger sample sizes incur lower costs.
- Genetic Research: TaqMan SNP genotyping is applicable across various realms of genetic studies involving SNP analysis, encompassing fields such as human genetics and the genetic breeding of flora and fauna.
- Whole-genome SNP association studies: Particularly well-suited for validating initial positive loci obtained from comprehensive whole-genome SNP association studies. This approach aids in pinpointing SNPs associated with specific traits or diseases, thereby facilitating a comprehensive understanding of their genetic foundations.
- Validation of preliminary mutation sites from whole-genome sequencing: In the course of whole-genome sequencing studies, a substantial number of preliminary mutation sites are typically identified. TaqMan SNP genotyping serves as a valuable tool for further validation of these sites with large sample sizes, elucidating their frequencies within specific populations or species and their correlations with phenotypic traits.
TaqMan SNP Genotyping Workflow
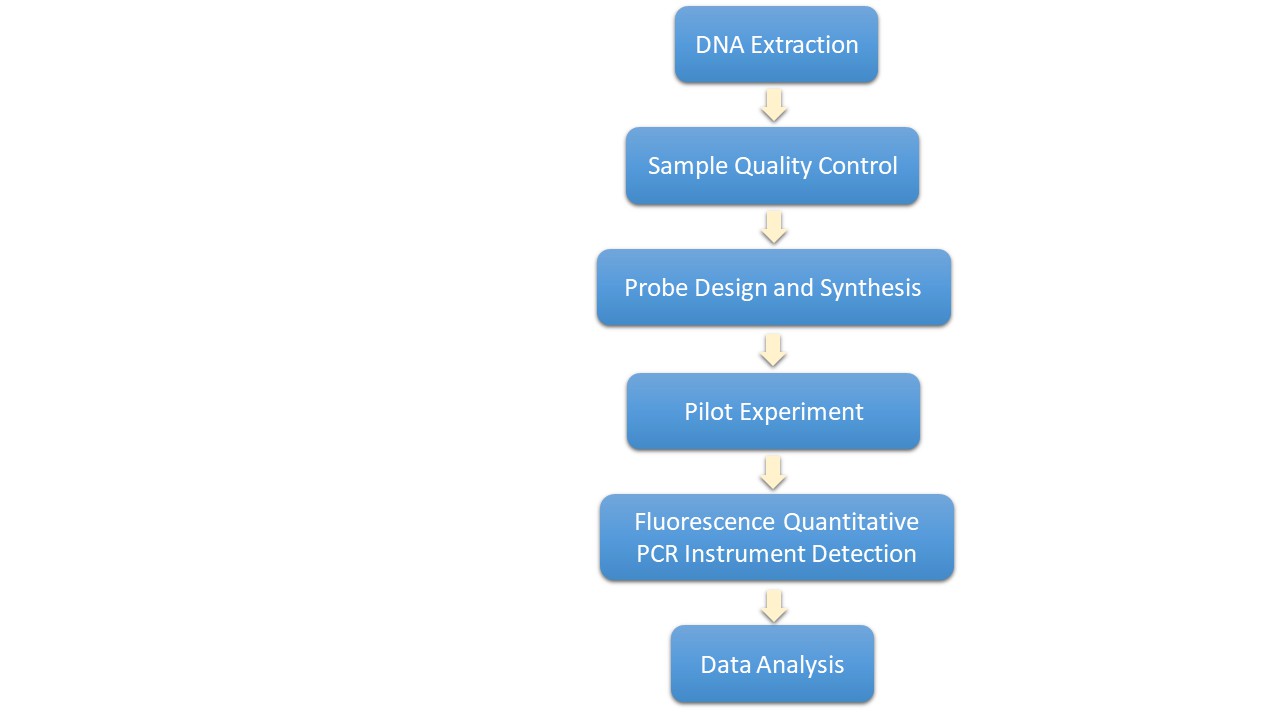
The experimental procedures encompass numerous stages including DNA extraction, sample quality inspection, probe design and synthesis, preliminary experiments, detection achieved through fluorescence quantitative PCR instruments, and subsequent data analysis. Central to the PCR reaction is the process of identifying the SNP genotypes present within the samples, accomplished via fluorescent signal detection.
Service Specifications
Sample requirements and preparation
|
|
Detection
|
|
Data Analysis
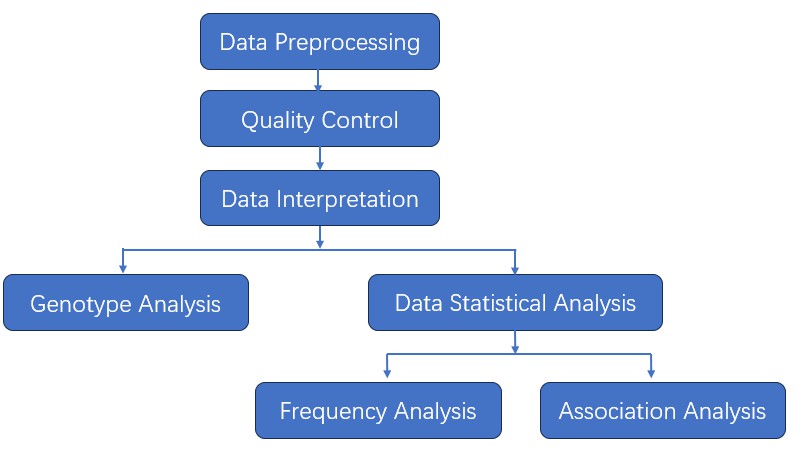
We provide multiple customized bioinformatics analyses:
|
Analysis Pipeline
Deliverables
Experimental conditions including information of primer and probe sequences, reagent names, device models, protocols, clustering graphs and genotyping tables of each SNP locus in individual sample.
CD genomics offers highly trusted and flexible TaqMan technology to meet your specific SNP genotyping needs.
Reference:
- Ziller M. J. et al. Targeted bisulfite sequencing of the dynamic DNA methylome. Epigenetics & chromatin, 2016, 9(1): 55.
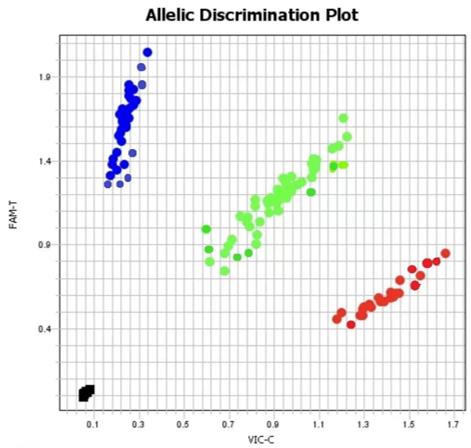 TaqMan probe-based genotyping detection results chart
TaqMan probe-based genotyping detection results chart
1. What is the working principle of TaqMan SNP gene typing?
This technology refers to the single nucleotide polymorphism (SNP) genotyping approach, meticulously developed by the ABI research team. The operating principle has been expounded as follows:
During the PCR reaction, a pair of Minor Groove Binder (MGB) specific probes, with unique fluorescent tags at both ends, is included to discern different allelic genes (allele1 and allele2). The 5' end carries a reporter fluorescent moiety, while the 3' end is equipped with a quenching fluorescent moiety. Throughout the PCR process, both probes have the capability to selectively anneal and bond with the complementary sequences located between the forward and reverse primers.
When these probes subsist in their intact state, the energy resonance transfer instigates the fluorescent moieties to emit a faint fluorescence. Once the specific probe bonds exclusively to its corresponding allelic gene, the DNA polymerase exerts its 5' to 3' exonuclease activity; cleaving the reporter fluorescent moiety and liberating it from the quenching effect of the 3' end quenching fluorescent moieties—this results in fluorescence emission.
The two probes are distinctly marked: the 5' end carries diverse fluorescence (either FAM or VIC), while the 3' end is attached to an MGB quenching moiety complex. Variances in detected fluorescences can thus be utilized to infer the SNP allelic genotype of the respective samples.
2. What are the advantages of TaqMan SNP genotyping?
TaqMan SNP genotyping offers simplicity, avoiding cross-contamination during post-PCR handling; real-time detection of SNP loci, providing rapid results; minimal sample requirement, suitable for limited samples; simultaneous analysis of large batches of samples, enhancing detection efficiency and throughput.
3. In which research areas is TaqMan SNP genotyping applicable?
TaqMan SNP genotyping is applicable to various genetic research fields involving SNP genotyping, including human genetics, and animal and plant breeding. It is particularly suitable for further validation studies of initial positive loci obtained from whole-genome SNP association studies, and a large number of preliminary screening mutation loci obtained from whole-genome sequencing.
4. What does TaqMan SNP genotyping contribute to clinical diagnosis?
The practical applications of TaqMan SNP genotyping within the realm of clinical diagnosis are significant. Through the effective utilization of this process, clinicians are empowered to uncover genetic variations specific to certain diseases. Subsequently, these insights are used to thoughtfully devise treatment plans personalized to the needs of individual patients. Moreover, TaqMan SNP genotyping can potentially enable the creation of proactive preventive measures, thus opening doors to preventative medicine.
5. What are the expediencies in precluding false positive results in TaqMan SNP genotyping experiments?
The matter of false positive occurrences in TaqMan SNP genotyping experiments is one of considerable attention. It is crucial to enforce diverse control measures in order to increase the precision and reliability of the yielded findings. Examples of implementable checks and controls include encompassing negative control samples and the execution of numerous reproducible experiments. Such strategies serve to reduce the incidence of inaccuracies within the research outcomes.
6. How does TaqMan SNP genotyping compare to other genotyping methods?
In comparison to alternative SNP genotyping techniques, TaqMan SNP genotyping offers heightened sensitivity, specificity, and expediency, coupled with ease of implementation. Consequently, it has garnered widespread adoption in practical applications.
7. How should failures in TaqMan SNP genotyping experiments be addressed?
In instances of experimental failure, a systematic approach is warranted. This includes a meticulous review of experimental procedures to ensure procedural accuracy, assessment of the freshness and quality of reagents, scrutiny for potential contamination and technical issues, and exploration of adjustments to experimental conditions aimed at enhancing outcomes.
SNP genotyping using TaqMan® technology: the CYP2D6*17 assay conundrum
Journal: Scientific reports
Impact factor: 4.746
Published: 19 March 2015
Background
CYP2D6 is a critical enzyme in drug metabolism, involved in metabolizing a significant portion of clinically used drugs. Its activity varies widely among populations, influencing medication efficacy and adverse events. Over 100 allelic variants have been identified, impacting drug metabolism phenotypes. TaqMan assays are commonly used for CYP2D6 genotyping, but challenges persist due to gene complexity. Unexpected findings in TaqMan assays prompted further investigation into the accuracy of genotype calls, highlighting the need for rigorous assay validation and understanding genotype-phenotype correlations for personalized medicine.
Methods
- DNA samples
- Two liver tissue samples
- Three DNA samples from the Coriell Institute
- CYP2D6 genotyping
- TaqMan assays
- Genotyping with RFLP
- Gene re-sequencing
- Secondary structure analysis
Results
TaqMan assays were used to genotype cases for CYP2D6 variants, revealing inconsistencies in calls for the 1023C>T SNP. Contamination and DNA quality issues were ruled out. Further investigation grouped subjects into those carrying CYP2D6*17 and 4 alleles. Cases exhibited homozygous calls for 1023C>T, later identified as carrying a CYP2D64 subvariant with a specific SNP trio. Others showed consistent calls or gene duplications. These findings highlight the complexity of CYP2D6 genotyping and the need for precise analysis methods.
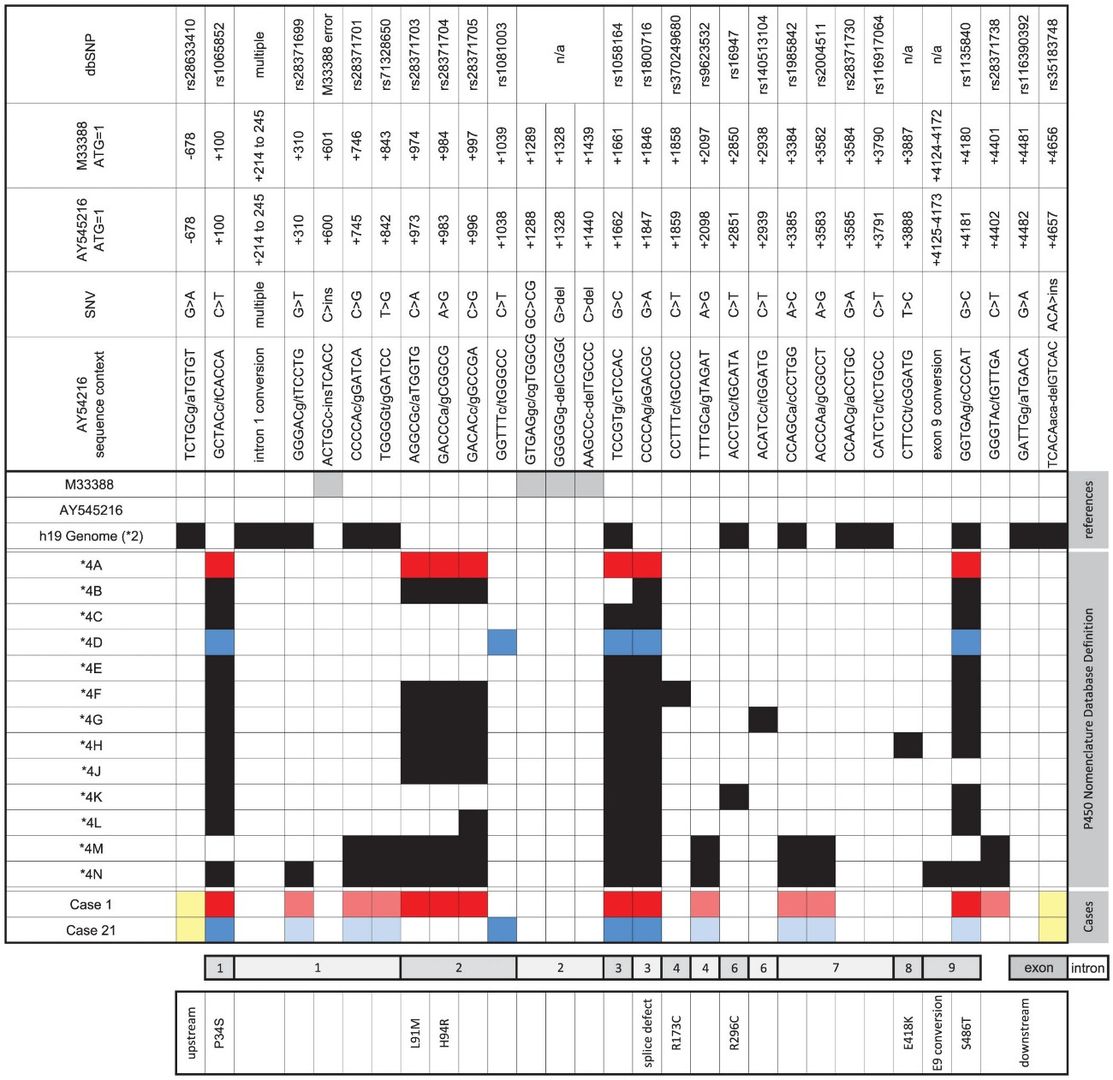 Figure 1. CYP2D6*4 subvariants.
Figure 1. CYP2D6*4 subvariants.
CYP2D6 genotyping across three institutions identified 16 subjects of African ancestry with heterozygous key SNPs but homozygous for 1023T/T. They were suggested to have a novel CYP2D6*4 subvariant carrying the 1023C>T SNP. However, RFLP analysis contradicted TaqMan results, indicating heterozygosity for 1023C>T. Gene resequencing confirmed RFLP findings, revealing common SNPs on CYP2D6*4 variants. This highlights the complexity of CYP2D6 genotyping and the need for comprehensive analysis methods.
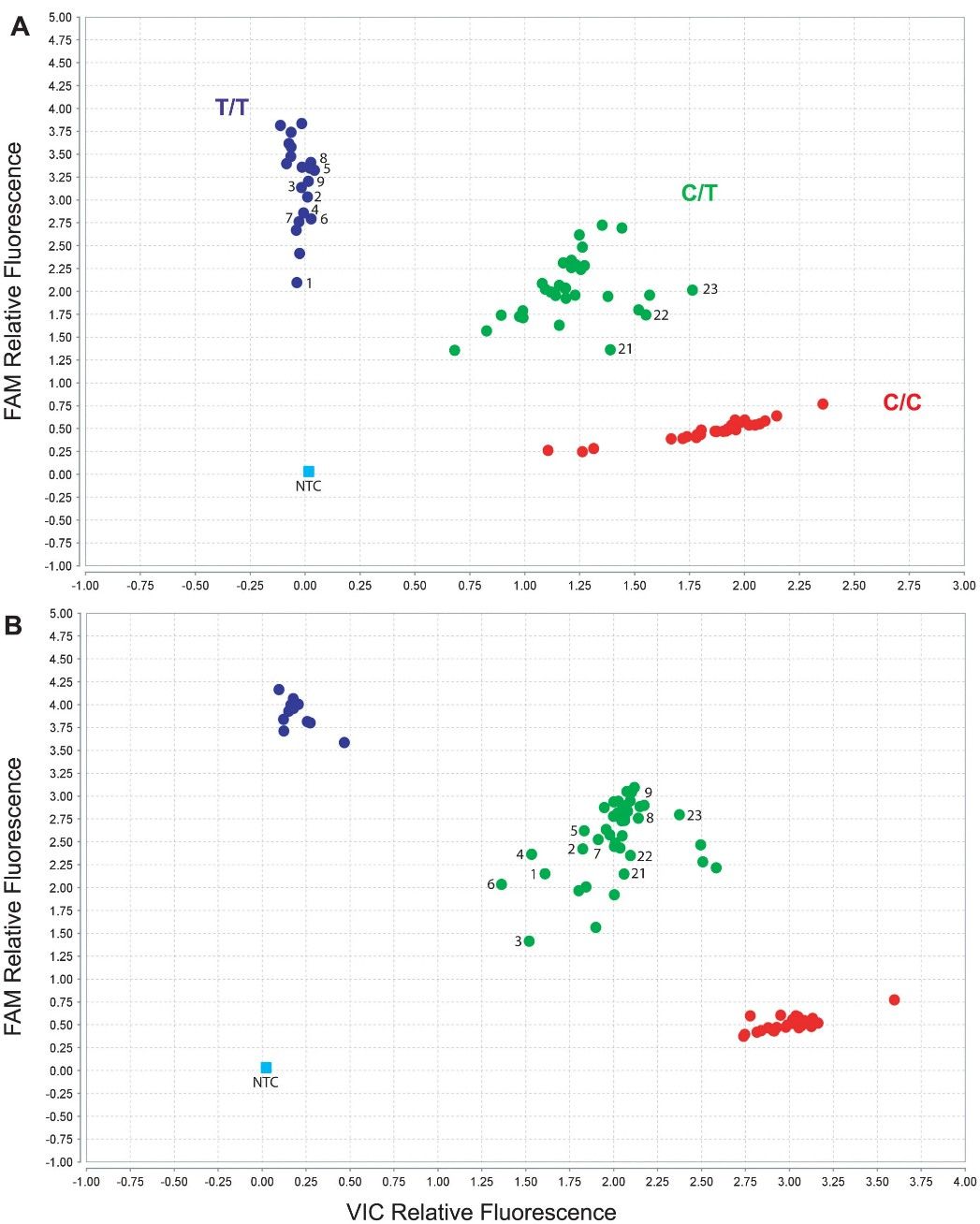 Figure 2. CYP2D6*17 (1023C>T) TaqMan Assay Genotype Results.
Figure 2. CYP2D6*17 (1023C>T) TaqMan Assay Genotype Results.
Collaboration with Thermo Fisher led to testing an alternative CYP2D6*17 assay design. This new design corrected misgenotyping observed in previous assays, identifying heterozygosity for CYP2D6*17 SNP in previously homozygous samples. Samples with CYP2D6*4/4 genotype were accurately called. Analysis of assay PCR product secondary structure suggested differences between CYP2D6*17 and *4 variants, potentially influencing assay performance.
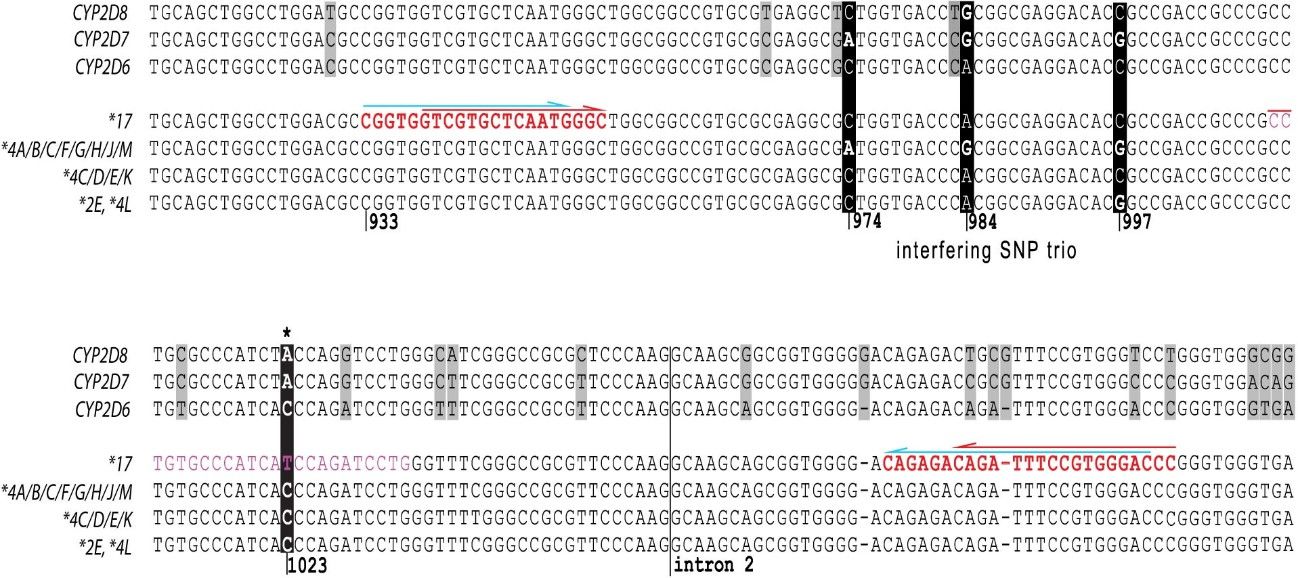 Figure 3. Sequence Comparison of CYP2D6 variants, CYP2D7 and CYP2D8
Figure 3. Sequence Comparison of CYP2D6 variants, CYP2D7 and CYP2D8
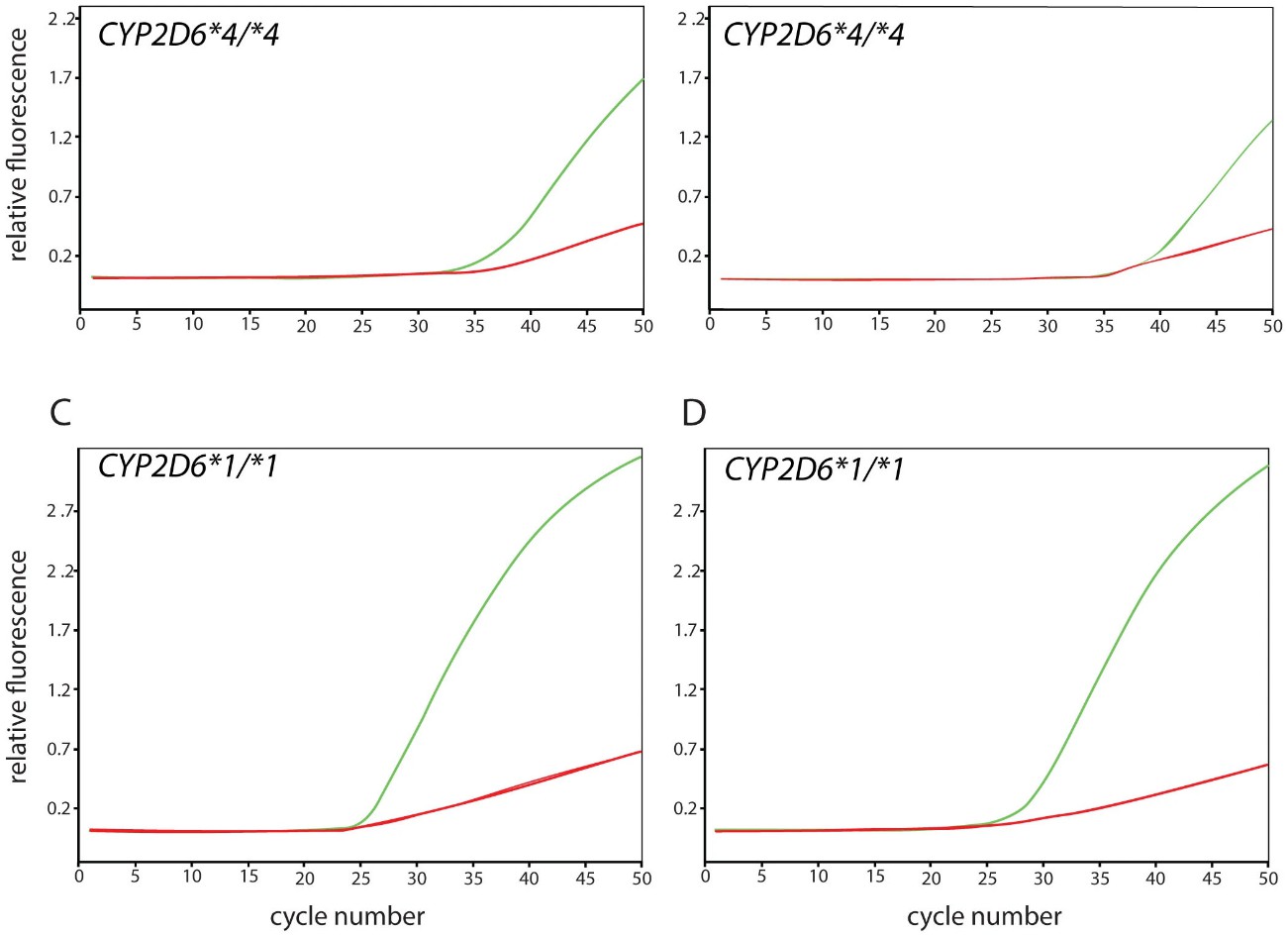 Figure 4. Real-time amplification for the original and alternate CYP2D6*17 TaqMan assays.
Figure 4. Real-time amplification for the original and alternate CYP2D6*17 TaqMan assays.
Conclusion
The mechanism behind allele drop-out remains elusive, possibly involving Taq polymerase properties or secondary structures. This unexpected phenomenon highlights the need for thorough assay validation and vigilance in interpreting results, particularly in highly polymorphic genes like CYP2D6.
Reference:
- Gaedigk A, Freeman N, Hartshorne T, et al. SNP genotyping using TaqMan® technology: the CYP2D6* 17 assay conundrum. Scientific reports, 2015, 5(1): 9257.
Here are some publications that have been successfully published using our services or other related services:
Investigating interactive effects of worry and the catechol-o-methyltransferase gene (COMT) on working memory performance
Journal: Cognitive, Affective, & Behavioral Neuroscience
Year: 2021
Eye color prediction using the IrisPlex system: a limited pilot study in the Iraqi population
Journal: Egyptian Journal of Forensic Sciences
Year: 2020
Embryonic origin and genetic basis of cave associated phenotypes in the isopod crustacean Asellus aquaticus
Journal: Scientific Reports
Year: 2023
Scanning indels in the 5q22.1 region and identification of the TMEM232 susceptibility gene that is associated with atopic dermatitis in the Chinese Han population
Journal: Gene
Year: 2017
Impact of the OATP1B1 c.521T>C single nucleotide polymorphism on the pharmacokinetics of exemestane in healthy post-menopausal female volunteers
Journal: Journal of clinical pharmacy and therapeutics
Year: 2017
See more articles published by our clients.


 Sample Submission Guidelines
Sample Submission Guidelines
