CD Genomics offers CGH microarray services for numerous species, including human, mouse, rat, and chicken, with custom CGH arrays available for additional species.
Their microarray CGH enables comprehensive genome-wide screening with high resolution, facilitating the detection of previously undetectable copy number imbalances such as deletions and duplications. By leveraging validated procedures and optimized protocols, along with high-resolution microarray scanning and sophisticated analytic software, CD Genomics ensures superior data quality and rapid turn-around times.
Introduction of CGH Microarray
Comparative Genomic Hybridization (CGH) microarray is a cutting-edge molecular cytogenetic tool designed to map and quantify variations in DNA copy number throughout the genome. This advanced technique is crucial for detecting chromosomal abnormalities linked to a broad spectrum of genetic disorders and malignancies.
Principles of CGH Microarray
CGH microarray operates based on the principle of comparative genomic hybridization. This involves labeling DNA samples from two sources—one from a test specimen, such as tumor DNA, and another from a reference sample, typically normal tissue—with distinct fluorescent dyes. These labeled DNA samples are then hybridized onto a microarray chip that is dotted with a grid of DNA probes representing various genomic regions.
The process starts with labeling DNA extracted from both the test and reference samples. The labeled DNAs are combined and applied to the microarray, where each probe interacts with its corresponding DNA sequence in the sample. The microarray is then scanned to measure the intensity of the fluorescent signals. By comparing the fluorescence of the test sample to that of the reference, researchers can determine gains or losses in specific chromosomal regions.
CGH microarray offers a high-resolution view of chromosomal changes, significantly surpassing traditional karyotyping in both sensitivity and resolution. While karyotyping is effective for detecting large-scale chromosomal aberrations, CGH microarray can identify submicroscopic alterations, such as microdeletions and microduplications. This precision is essential for diagnosing complex genetic conditions and cancers that might elude traditional diagnostic methods.
What does CGH Microarray Test For
CGH microarray is primarily utilized to detect genomic imbalances, including:
- Copy Number Variations (CNVs): These are deletions or duplications of DNA segments that can contribute to genetic disorders. CNVs can be further classified into microdeletions and microduplications, with sizes ranging from a few kilobases to several megabases.
- Chromosomal Rearrangements: This technique can identify structural variations across the genome, encompassing deletions, duplications, and complex rearrangements.
- Genomic Imbalances: CGH microarray is proficient in detecting imbalances associated with various conditions, including cancers, genetic syndromes, and congenital anomalies.
However, it is important to note that despite its comprehensive capabilities, CGH microarray cannot detect balanced chromosomal rearrangements such as translocations or inversions, which do not change the overall DNA copy number but may still impact gene function.
Advantages of CGH Microarray Service
- High Resolution and Sensitivity: Detects chromosomal imbalances as small as 10 kilobases, exceeding the resolution of traditional karyotyping.
- Comprehensive Genomic Coverage: Analyzes the entire genome to identify previously undetected imbalances and novel CNVs.
- Faster Turnaround Time: Provides quicker results by analyzing thousands of genomic regions simultaneously.
- Improved Detection of Submicroscopic Aberrations: Identifies tiny chromosomal changes often missed by other methods.
- Reduced Need for Sample Preprocessing: Minimizes contamination risks with less extensive sample preparation.
CGH Microarray Workflow
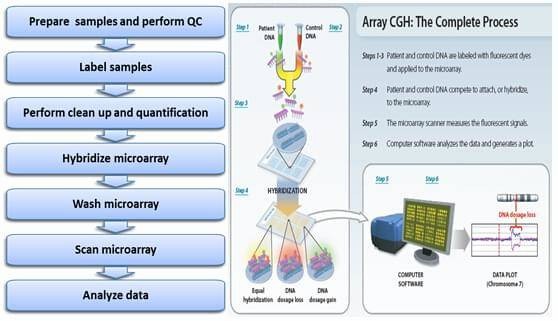
The CGH microarray workflow encompasses several critical steps. Initially, high-quality genomic DNA is extracted from biological samples and labeled with fluorescent dyes, typically Cy3 for the experimental sample and Cy5 for the reference sample. This labeled DNA is subsequently hybridized onto a high-density CGH microarray chip, which comprises a grid of DNA probes.
Following the hybridization process, the arrays are washed to remove non-specifically bound DNA. The next step involves scanning the arrays with a high-resolution scanner to detect fluorescent signals. Specialized software is then employed to analyze these signals, determining the relative copy number of genomic regions. By comparing the experimental sample to the reference sample, areas of genomic gain or loss are identified.
Service Specifications
Sample Requirements
|
|
Click |
Sequencing Strategy
|
| Bioinformatics Analysis We provide multiple customized bioinformatics analyses:
|
Recommendations and Custom Service
Table 1 Agilent CGH microarrays
| Organism | Array format(s) |
|---|---|
| Human | 4x44k, 8x60k, 2x105k, 4x180k,1x244k, 2x400k, 1x1M |
| Mouse | 2x105k, 4x180k, 1x244k,1x1M |
| Rat | 2x105k, 4x180k, 1x244k,1x1M |
| Others (please click here) | Please enquire |
Analysis Pipeline
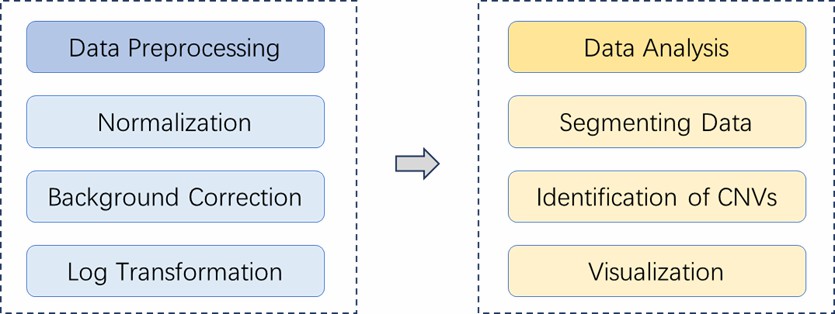
Deliverables
- Raw data
- Segmentation analysis based on the DNA copy package
- Segmentation GFF file
- Genome annotation GFF file
CD Genomics can also help create your own custom CGH microarray. We are ready to help you with your custom array needs, whether it's a standard design or something more creative. CGH microarray services offered by CD Genomics provide a state-of-the-art approach to genomic analysis, combining high resolution, comprehensive coverage, and fast turnaround times to meet the needs of research applications.
For details, please feel free to contact us with any questions by completing a quote request.
Partial results are shown below:
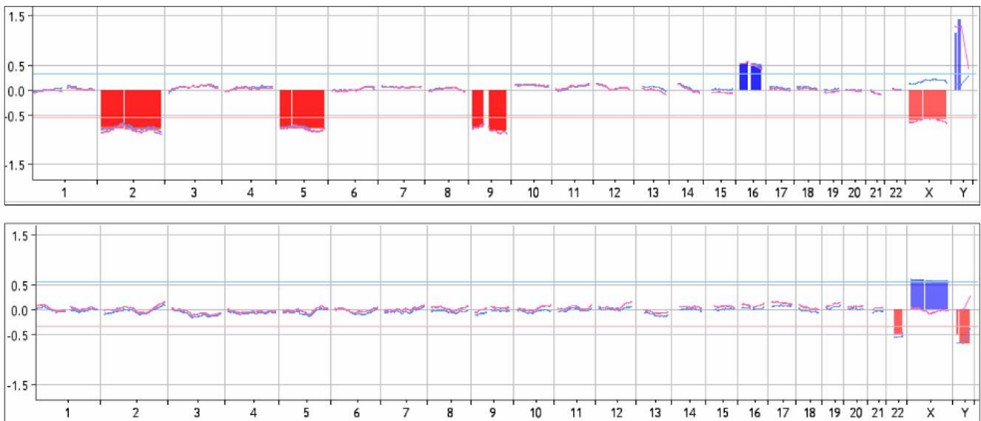 CNV Detection Results Diagram
CNV Detection Results Diagram
1. What is the difference between CGH microarray and SNP microarray?
CGH microarray focuses on detecting CNVs and genomic imbalances by comparing fluorescent signals from the experimental and reference samples. In contrast, SNP microarray detects single nucleotide polymorphisms and can identify both CNVs and allele-specific imbalances.
2. Can CGH microarray detect balanced chromosomal rearrangements?
No, CGH microarray cannot detect balanced chromosomal rearrangements such as translocations or inversions. To identify these types of chromosomal changes, additional techniques like spectral karyotyping or fluorescence in situ hybridization (FISH) are required.
3. How reliable is CGH microarray for clinical use?
CGH microarray is highly reliable for detecting genomic imbalances and is widely used in clinical settings. Its high resolution and comprehensive coverage make it a valuable tool for diagnosing genetic disorders.
4. What are the limitations of CGH microarray?
The main limitations of CGH microarray include its inability to detect balanced chromosomal rearrangements and the potential for identifying CNVs of uncertain clinical significance. Interpretation of some CNVs may require further validation.
The Use of CGH Arrays for Identifying Copy Number Variations in Children with Autism Spectrum Disorder
Journal: Brain Sciences
Impact factor: 3.333
Published: 22 May 2024
Background
Autism Spectrum Disorder (ASD) is a common neurodevelopmental disorder affecting social interaction, communication, and behavior, with a higher prevalence in males. It is usually diagnosed between ages 2 and 4. ASD often co-occurs with conditions like intellectual disabilities and epilepsy. Genetic factors, including syndromes like Fragile X, play a significant role, with CNVs contributing to about 10% of cases. CGH microarray is an effective tool for detecting these genetic variations, offering higher resolution and accuracy than traditional karyotyping.
Materials & Methods
Sample Preparation:
- Patients with ASD
- Peripheral venous blood
- DNA extraction
Method:
- aCGH
- FISH
- Mapped to the reference genome
- Identification of CNVs
Results
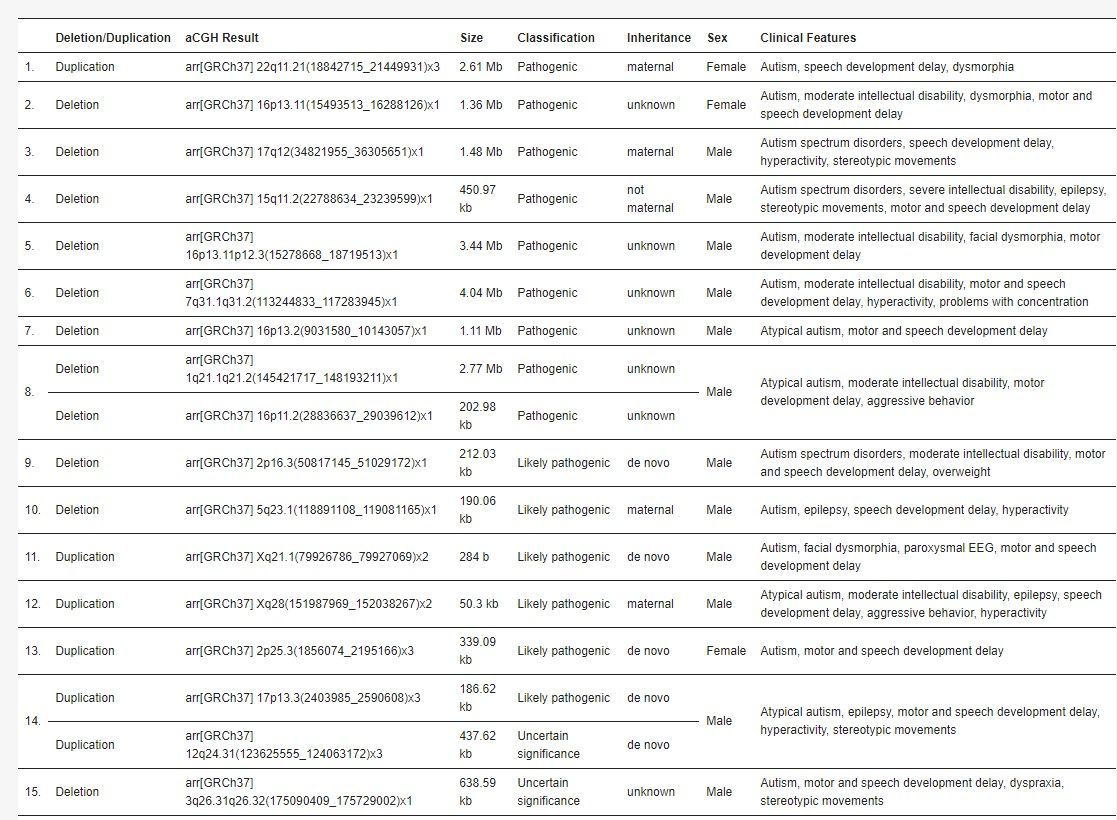
Variants were categorized based on ACMG guidelines into pathogenic, likely pathogenic, VUS, likely benign, and benign. In 14 out of 180 patients (7.8%), nine pathogenic and six likely pathogenic variants were found. There were also 20 VUSs and five likely benign variants. Pathogenic variants were more common in males and included deletions and duplications ranging from 284 bp to 4.04 Mb. In patients with isolated ASD, two pathogenic and one likely pathogenic variants were identified, while those with additional features had more pathogenic variants. Parental diagnostics revealed that many variants were de novo, though some were inherited. The study used a specialized microarray that detected more imbalances than standard arrays. Identified variants include several associated with known syndromes and ASD, while some VUSs require further investigation.
 Figure 1. The number of variants identified in each pathogenicity category in patients with isolated ASD (a) and ASD with additional clinical features (b).
Figure 1. The number of variants identified in each pathogenicity category in patients with isolated ASD (a) and ASD with additional clinical features (b).
Table 1. A detailed list of pathogenic and likely pathogenic variants, including aCGH result, size of CNV, inheritance (if known), sex, and clinical phenotype.
Conclusion
Standard microarrays may lack resolution for accurate ASD assessment, so future research should use high-resolution tools and combine whole exome sequencing with CNV analysis. Reporting VUSs remains crucial as their significance is clarified, aiding the understanding of genotype-phenotype correlations in ASD.
Reference:
- Kucińska A, Hawuła W, Rutkowska L, et al. The Use of CGH Arrays for Identifying Copy Number Variations in Children with Autism Spectrum Disorder. Brain Sciences. 2024 Mar 13;14(3):273.


 Sample Submission Guidelines
Sample Submission Guidelines
